Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004301A_C01 KMC004301A_c01
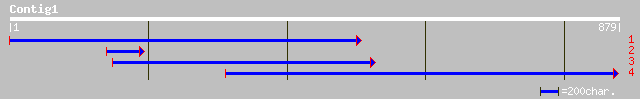
(879 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_176374.1| glycosyl hydrolase family 1; protein id: At1g61... 234 1e-60
ref|NP_176375.2| glycosyl hydrolase family 1; protein id: At1g61... 230 2e-59
pir||T02128 beta-glucosidase homolog F8K4.3 - Arabidopsis thalia... 230 2e-59
ref|NP_193907.2| glycosyl hydrolase family 1; protein id: At4g21... 224 1e-57
gb|AAC69619.1| beta-glucosidase [Pinus contorta] 161 1e-38
>ref|NP_176374.1| glycosyl hydrolase family 1; protein id: At1g61810.1 [Arabidopsis
thaliana] gi|7435449|pir||T02127 beta-glucosidase
homolog F8K4.2 - Arabidopsis thaliana
gi|3367516|gb|AAC28501.1| Similar to beta-glucosidase
BGQ60 precursor gb|L41869 from Hordeum vulgare.
[Arabidopsis thaliana]
Length = 520
Score = 234 bits (597), Expect = 1e-60
Identities = 104/202 (51%), Positives = 144/202 (70%)
Frame = -1
Query: 879 PAEMEEILGPDLPPFSRYDVENKLKRGLDFIGVNHYTSMFVKDCIFSACEAGLGSSRTEG 700
P EM +ILGP LP FS +V+N K DF+G+NHYTS F++DC+ SAC G G+ + EG
Sbjct: 309 PKEMVDILGPALPQFSSNEVKNLEKSRADFVGINHYTSYFIQDCLTSACNTGHGAFKAEG 368
Query: 699 FAFISGQMDGVSIGEPTVLDWLHVHPQGMEKIVTYIKDRYNNIPMFITENGLGTNENSFP 520
+A + V+IGE T ++W H+ P G K++ Y+KDRY N+PMFITENG G +
Sbjct: 369 YALKLDRKGNVTIGELTDVNWQHIDPTGFHKMLNYLKDRYPNMPMFITENGFGDLQKPET 428
Query: 519 TTEDIFNDADRVEYLSGYLDSLASAIRKGADARGYFVWSLLDNFEWIQGYTIRFGLHHLD 340
T +++ ND R++Y+SGYL++L +A+R GA+ +GYFVWSLLDNFEW+ GY +RFGL H+D
Sbjct: 429 TDKELLNDTKRIQYMSGYLEALQAAMRDGANVKGYFVWSLLDNFEWLFGYKVRFGLFHVD 488
Query: 339 YTTLSRTRRLSAFWYRNFIARH 274
TTL R+ + SA WY+N+I H
Sbjct: 489 LTTLKRSPKQSASWYKNYIEEH 510
>ref|NP_176375.2| glycosyl hydrolase family 1; protein id: At1g61820.1 [Arabidopsis
thaliana]
Length = 516
Score = 230 bits (587), Expect = 2e-59
Identities = 101/202 (50%), Positives = 145/202 (71%)
Frame = -1
Query: 879 PAEMEEILGPDLPPFSRYDVENKLKRGLDFIGVNHYTSMFVKDCIFSACEAGLGSSRTEG 700
P EM +LG LP FS ++ + + DF+G+NHYTS F++DC+ +AC +G G+S++EG
Sbjct: 306 PEEMVNLLGSALPKFSSNEMNSLMSYKSDFLGINHYTSYFIQDCLITACNSGDGASKSEG 365
Query: 699 FAFISGQMDGVSIGEPTVLDWLHVHPQGMEKIVTYIKDRYNNIPMFITENGLGTNENSFP 520
A + VSIGE T ++W H+ P G K++ Y+K+RY+NIPM+ITENG G +
Sbjct: 366 LALKLDRKGNVSIGELTDVNWQHIDPNGFRKMLNYLKNRYHNIPMYITENGFGQLQKPET 425
Query: 519 TTEDIFNDADRVEYLSGYLDSLASAIRKGADARGYFVWSLLDNFEWIQGYTIRFGLHHLD 340
T E++ +D R++YLSGYLD+L +A+R GA+ +GYF WSLLDNFEW+ GY +RFGL H+D
Sbjct: 426 TVEELLHDTKRIQYLSGYLDALKAAMRDGANVKGYFAWSLLDNFEWLYGYKVRFGLFHVD 485
Query: 339 YTTLSRTRRLSAFWYRNFIARH 274
+TTL RT + SA WY+NFI ++
Sbjct: 486 FTTLKRTPKQSATWYKNFIEQN 507
>pir||T02128 beta-glucosidase homolog F8K4.3 - Arabidopsis thaliana
gi|3367517|gb|AAC28502.1| Similar to F4I1.26 putative
beta-glucosidase gi|3128187 from A. thaliana BAC
gb|AC004521. ESTs gb|N97083, gb|F19868 and gb|F15482
come from this gene. [Arabidopsis thaliana]
Length = 527
Score = 230 bits (587), Expect = 2e-59
Identities = 101/202 (50%), Positives = 145/202 (71%)
Frame = -1
Query: 879 PAEMEEILGPDLPPFSRYDVENKLKRGLDFIGVNHYTSMFVKDCIFSACEAGLGSSRTEG 700
P EM +LG LP FS ++ + + DF+G+NHYTS F++DC+ +AC +G G+S++EG
Sbjct: 317 PEEMVNLLGSALPKFSSNEMNSLMSYKSDFLGINHYTSYFIQDCLITACNSGDGASKSEG 376
Query: 699 FAFISGQMDGVSIGEPTVLDWLHVHPQGMEKIVTYIKDRYNNIPMFITENGLGTNENSFP 520
A + VSIGE T ++W H+ P G K++ Y+K+RY+NIPM+ITENG G +
Sbjct: 377 LALKLDRKGNVSIGELTDVNWQHIDPNGFRKMLNYLKNRYHNIPMYITENGFGQLQKPET 436
Query: 519 TTEDIFNDADRVEYLSGYLDSLASAIRKGADARGYFVWSLLDNFEWIQGYTIRFGLHHLD 340
T E++ +D R++YLSGYLD+L +A+R GA+ +GYF WSLLDNFEW+ GY +RFGL H+D
Sbjct: 437 TVEELLHDTKRIQYLSGYLDALKAAMRDGANVKGYFAWSLLDNFEWLYGYKVRFGLFHVD 496
Query: 339 YTTLSRTRRLSAFWYRNFIARH 274
+TTL RT + SA WY+NFI ++
Sbjct: 497 FTTLKRTPKQSATWYKNFIEQN 518
>ref|NP_193907.2| glycosyl hydrolase family 1; protein id: At4g21760.1 [Arabidopsis
thaliana]
Length = 535
Score = 224 bits (571), Expect = 1e-57
Identities = 107/204 (52%), Positives = 140/204 (68%)
Frame = -1
Query: 879 PAEMEEILGPDLPPFSRYDVENKLKRGLDFIGVNHYTSMFVKDCIFSACEAGLGSSRTEG 700
P EM EILG DLP F++ D+++ K LDFIG+N YTS + KDC+ S CE G G SR EG
Sbjct: 329 PREMREILGDDLPEFTKDDLKSS-KNALDFIGINQYTSRYAKDCLHSVCEPGKGGSRAEG 387
Query: 699 FAFISGQMDGVSIGEPTVLDWLHVHPQGMEKIVTYIKDRYNNIPMFITENGLGTNENSFP 520
F + + DG+ +GEP GME+++ Y +RY NI +++TENG G N
Sbjct: 388 FVYANALKDGLRLGEPV----------GMEEMLMYATERYKNITLYVTENGFGENN---- 433
Query: 519 TTEDIFNDADRVEYLSGYLDSLASAIRKGADARGYFVWSLLDNFEWIQGYTIRFGLHHLD 340
T + ND RV+++S YLD+L A+RKGAD RGYF WSLLDNFEWI GYTIRFG++H+D
Sbjct: 434 -TGVLLNDYQRVKFMSNYLDALKRAMRKGADVRGYFAWSLLDNFEWISGYTIRFGMYHVD 492
Query: 339 YTTLSRTRRLSAFWYRNFIARHKA 268
++T RT RLSA WY+NFI +H+A
Sbjct: 493 FSTQERTPRLSASWYKNFIFQHRA 516
>gb|AAC69619.1| beta-glucosidase [Pinus contorta]
Length = 513
Score = 161 bits (408), Expect = 1e-38
Identities = 89/215 (41%), Positives = 128/215 (59%), Gaps = 6/215 (2%)
Frame = -1
Query: 879 PAEMEEILGPDLPPFSRYDVENKLKRGLDFIGVNHYTSMFVKDCIFSACEAGLGSSRTEG 700
P EM E LG LP S ++ KL+ D++G+NHYT+++ + L T+
Sbjct: 302 PQEMRERLGSRLPSISS-ELSAKLRGSFDYMGINHYTTLY------ATSTPPLSPDHTQY 354
Query: 699 F-----AFISGQMDGVSIGEPTVLDWLHVHPQGMEKIVTYIKDRYNNIPMFITENGLGTN 535
+++G+ GVSIGE T +D L V P G++KIV Y+K+ Y+N + I ENG +
Sbjct: 355 LYPDSRVYLTGERHGVSIGERTGMDGLFVVPHGIQKIVEYVKEFYDNPTIIIAENGYPES 414
Query: 534 ENSFPTTEDIFNDADRVEYLSGYLDSLASAIRKGADARGYFVWSLLDNFEWIQGYTIRFG 355
E S T ++ ND R+ + L L++AI+ G+D RGYFVWSLLDNFEW GYTIRFG
Sbjct: 415 EESSSTLQENLNDVRRIRFHGDCLSYLSAAIKNGSDVRGYFVWSLLDNFEWAFGYTIRFG 474
Query: 354 LHHLDY-TTLSRTRRLSAFWYRNFIARHKASAGIR 253
L+H+D+ + R +LSA W+R F+ +H IR
Sbjct: 475 LYHVDFISDQKRYPKLSAQWFRQFL-QHDDQGSIR 508
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 776,805,800
Number of Sequences: 1393205
Number of extensions: 17800414
Number of successful extensions: 47073
Number of sequences better than 10.0: 525
Number of HSP's better than 10.0 without gapping: 44198
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46324
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 47382100290
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)