Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
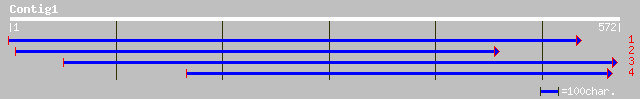
Query= KMC004296A_C01 KMC004296A_c01
(566 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|O04161|AT12_LYCES Ammonium transporter 1, member 2 (LeAMT1;2)... 91 1e-17
gb|AAD54639.1|AF083036_1 ammonium transporter [Arabidopsis thali... 89 3e-17
ref|NP_176658.1| ammonium transporter, puitative; protein id: At... 89 3e-17
gb|AAG24944.1| putative ammonium transporter AMT1;1 [Lotus japon... 81 8e-15
emb|CAC10555.1| ammonium transporter (AMT1.1) [Lotus japonicus] 81 8e-15
>sp|O04161|AT12_LYCES Ammonium transporter 1, member 2 (LeAMT1;2) gi|7450343|pir||T06585
ammonium transporter 2 - tomato
gi|2065194|emb|CAA64475.1| ammonium transporter
[Lycopersicon esculentum]
Length = 514
Score = 90.5 bits (223), Expect = 1e-17
Identities = 44/65 (67%), Positives = 50/65 (76%), Gaps = 1/65 (1%)
Frame = -2
Query: 565 GPLFYGLHKMKLLRISEDDETAGMDLTRHGGFAYAYHDEDDVSTKRGVMMSRIGP-GSSS 389
GPLFY LHK KLLRIS DDETAGMDLTRHGGFAY YHDED+ S+ G M+R+ P +S+
Sbjct: 443 GPLFYLLHKFKLLRISRDDETAGMDLTRHGGFAYIYHDEDEGSSMPGFKMTRVEPTNTST 502
Query: 388 PSTMN 374
P N
Sbjct: 503 PDHQN 507
>gb|AAD54639.1|AF083036_1 ammonium transporter [Arabidopsis thaliana]
Length = 514
Score = 89.4 bits (220), Expect = 3e-17
Identities = 45/66 (68%), Positives = 50/66 (75%), Gaps = 1/66 (1%)
Frame = -2
Query: 565 GPLFYGLHKMKLLRISEDDETAGMDLTRHGGFAYAYHDEDDVSTKR-GVMMSRIGPGSSS 389
GPLFYGLHKM LLRIS +DE AGMD+TRHGGFAYAY+DEDDVSTK G R+ P S S
Sbjct: 446 GPLFYGLHKMNLLRISAEDEMAGMDMTRHGGFAYAYNDEDDVSTKPWGHFAGRVEPTSRS 505
Query: 388 PSTMNT 371
+ T
Sbjct: 506 STPTPT 511
>ref|NP_176658.1| ammonium transporter, puitative; protein id: At1g64780.1, supported
by cDNA: gi_17064989, supported by cDNA: gi_20260051,
supported by cDNA: gi_4324713, supported by cDNA:
gi_5880356 [Arabidopsis thaliana]
gi|22001525|sp|Q9ZPJ8|AT12_ARATH Ammonium transporter 1,
member 2 (AtAMT1;2) gi|25327164|pir||A96671 Ammonium
transporter ATM1,2 [imported] - Arabidopsis thaliana
gi|4324714|gb|AAD17001.1| ammonium transporter
[Arabidopsis thaliana]
gi|5042414|gb|AAD38253.1|AC006193_9 Ammonium transporter
ATM1;2 [Arabidopsis thaliana] gi|17064990|gb|AAL32649.1|
Ammonium transporter ATM1 [Arabidopsis thaliana]
gi|20260052|gb|AAM13373.1| ammonium transporter ATM1;2
[Arabidopsis thaliana]
Length = 514
Score = 89.4 bits (220), Expect = 3e-17
Identities = 45/66 (68%), Positives = 50/66 (75%), Gaps = 1/66 (1%)
Frame = -2
Query: 565 GPLFYGLHKMKLLRISEDDETAGMDLTRHGGFAYAYHDEDDVSTKR-GVMMSRIGPGSSS 389
GPLFYGLHKM LLRIS +DE AGMD+TRHGGFAYAY+DEDDVSTK G R+ P S S
Sbjct: 446 GPLFYGLHKMNLLRISAEDEMAGMDMTRHGGFAYAYNDEDDVSTKPWGHFAGRVEPTSRS 505
Query: 388 PSTMNT 371
+ T
Sbjct: 506 STPTPT 511
>gb|AAG24944.1| putative ammonium transporter AMT1;1 [Lotus japonicus]
Length = 501
Score = 81.3 bits (199), Expect = 8e-15
Identities = 44/70 (62%), Positives = 52/70 (73%)
Frame = -2
Query: 565 GPLFYGLHKMKLLRISEDDETAGMDLTRHGGFAYAYHDEDDVSTKRGVMMSRIGPGSSSP 386
GPLF+ L+KMKLLRIS +DE AGMDLTRHGGFAYAY EDD S K G+ + +I P SSS
Sbjct: 439 GPLFFILNKMKLLRISTEDELAGMDLTRHGGFAYAY--EDDESHKPGIQLRKIEPNSSS- 495
Query: 385 STMNTPAASA 356
TP+A +
Sbjct: 496 ----TPSAES 501
>emb|CAC10555.1| ammonium transporter (AMT1.1) [Lotus japonicus]
Length = 502
Score = 81.3 bits (199), Expect = 8e-15
Identities = 44/70 (62%), Positives = 52/70 (73%)
Frame = -2
Query: 565 GPLFYGLHKMKLLRISEDDETAGMDLTRHGGFAYAYHDEDDVSTKRGVMMSRIGPGSSSP 386
GPLF+ L+KMKLLRIS +DE AGMDLTRHGGFAYAY EDD S K G+ + +I P SSS
Sbjct: 440 GPLFFILNKMKLLRISTEDELAGMDLTRHGGFAYAY--EDDESHKPGIQLRKIEPNSSS- 496
Query: 385 STMNTPAASA 356
TP+A +
Sbjct: 497 ----TPSAES 502
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 532,078,627
Number of Sequences: 1393205
Number of extensions: 12731685
Number of successful extensions: 39890
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 34773
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38854
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)