Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004286A_C01 KMC004286A_c01
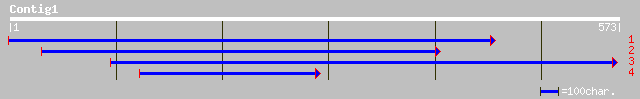
(572 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P52424|PUR5_VIGUN Phosphoribosylformylglycinamidine cyclo-lig... 79 5e-14
gb|AAO39864.1| putative 5'-phosphoribosyl-5-aminoimidazole synth... 65 6e-10
ref|NP_191061.1| phosphoribosylformylglycinamidine cyclo-ligase ... 64 1e-09
ref|ZP_00060728.1| hypothetical protein [Clostridium thermocellu... 46 4e-04
gb|EAA27164.1| hypothetical protein [Neurospora crassa] 46 4e-04
>sp|P52424|PUR5_VIGUN Phosphoribosylformylglycinamidine cyclo-ligase, chloroplast
precursor (AIRS) (Phosphoribosyl-aminoimidazole
synthetase) (AIR synthase) gi|7437995|pir||T10963
phosphoribosylformylglycinamidine cyclo-ligase (EC
6.3.3.1) - cowpea gi|945060|gb|AAC14578.1|
aminoimidazole ribonucleotide (AIRS) synthetase [Vigna
unguiculata]
Length = 388
Score = 78.6 bits (192), Expect = 5e-14
Identities = 37/46 (80%), Positives = 41/46 (88%)
Frame = -2
Query: 571 MRRTFNMGIGMVLVVSPEAANRILENRDDTEKAYRIGEVISGKGVT 434
MRRTFNMGIGM+LVVSPEAANRILEN+ +K YRIGE+ISG GVT
Sbjct: 341 MRRTFNMGIGMILVVSPEAANRILENKGQADKFYRIGEIISGNGVT 386
>gb|AAO39864.1| putative 5'-phosphoribosyl-5-aminoimidazole synthetase [Oryza
sativa (japonica cultivar-group)]
Length = 398
Score = 65.1 bits (157), Expect = 6e-10
Identities = 34/45 (75%), Positives = 36/45 (79%)
Frame = -2
Query: 571 MRRTFNMGIGMVLVVSPEAANRILENRDDTEKAYRIGEVISGKGV 437
MRRTFNMGIGMVLVVS EAA+ ILE AYRIGEVISG+GV
Sbjct: 351 MRRTFNMGIGMVLVVSKEAADGILEGTHGPNHAYRIGEVISGEGV 395
>ref|NP_191061.1| phosphoribosylformylglycinamidine cyclo-ligase precursor; protein
id: At3g55010.1, supported by cDNA: gi_16974614
[Arabidopsis thaliana] gi|21264504|sp|Q05728|PUR5_ARATH
Phosphoribosylformylglycinamidine cyclo-ligase,
chloroplast precursor (AIRS)
(Phosphoribosyl-aminoimidazole synthetase) (AIR
synthase) gi|11272444|pir||T47640
phosphoribosylformylglycinamidine cyclo-ligase precursor
- Arabidopsis thaliana gi|7329631|emb|CAB82696.1|
phosphoribosylformylglycinamidine cyclo-ligase precursor
[Arabidopsis thaliana] gi|16974615|gb|AAL31210.1|
AT3g55010/T15C9_10 [Arabidopsis thaliana]
gi|22655452|gb|AAM98318.1| At3g55010/T15C9_10
[Arabidopsis thaliana]
Length = 389
Score = 64.3 bits (155), Expect = 1e-09
Identities = 31/48 (64%), Positives = 42/48 (86%), Gaps = 1/48 (2%)
Frame = -2
Query: 571 MRRTFNMGIGMVLVVSPEAANRILEN-RDDTEKAYRIGEVISGKGVTH 431
MRRTFN+GIGMV+VVSPEAA+RILE ++ AYR+GEV++G+GV++
Sbjct: 341 MRRTFNLGIGMVMVVSPEAASRILEEVKNGDYVAYRVGEVVNGEGVSY 388
>ref|ZP_00060728.1| hypothetical protein [Clostridium thermocellum ATCC 27405]
Length = 340
Score = 45.8 bits (107), Expect = 4e-04
Identities = 24/44 (54%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Frame = -2
Query: 571 MRRTFNMGIGMVLVVSPEAANRILEN-RDDTEKAYRIGEVISGK 443
M TFNMGIGM + V E AN ++E D E+AY IGEV+S K
Sbjct: 291 MYNTFNMGIGMTIAVDAEIANSVVEYLNKDKEQAYIIGEVVSDK 334
>gb|EAA27164.1| hypothetical protein [Neurospora crassa]
Length = 789
Score = 45.8 bits (107), Expect = 4e-04
Identities = 21/45 (46%), Positives = 34/45 (74%), Gaps = 1/45 (2%)
Frame = -2
Query: 571 MRRTFNMGIGMVLVVSPEAANRILENRD-DTEKAYRIGEVISGKG 440
M RTFN G+GMVL V+PEAA+ +++ + + EK Y IG++++ +G
Sbjct: 728 MARTFNNGVGMVLAVAPEAADAVVKGLEAEGEKVYTIGKLVNREG 772
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,480,209
Number of Sequences: 1393205
Number of extensions: 9684680
Number of successful extensions: 22952
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 22407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22943
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)