Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004277A_C01 KMC004277A_c01
(776 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
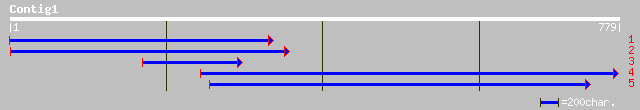
emb|CAA69931.1| PR10-1 protein [Medicago truncatula] gi|13928069... 165 6e-40
sp|Q43560|PR1_MEDSA Class-10 pathogenesis-related protein 1 (MSP... 164 1e-39
pir||T09526 stress response gene 1 - alfalfa gi|1136333|gb|AAB58... 162 5e-39
sp|P14710|DRR3_PEA Disease resistance response protein Pi49 (PR1... 161 9e-39
sp|P26987|SAM2_SOYBN Stress-induced protein SAM22 gi|99918|pir||... 161 9e-39
>emb|CAA69931.1| PR10-1 protein [Medicago truncatula] gi|13928069|emb|CAC37690.1|
class 10 PR protein [Medicago sativa]
Length = 157
Score = 165 bits (418), Expect = 6e-40
Identities = 77/111 (69%), Positives = 96/111 (86%)
Frame = -3
Query: 774 NGSAGTIKKITFEEDGQTKDLLHQIESIDEAHFGYSYSIIGGSDLPDPVEKISFEAQLVA 595
NG AGTIKK+TF E G+TK LH+++ +D+ +F Y+YSI+GG LPD VEKISFE++L A
Sbjct: 47 NGGAGTIKKLTFVEGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSA 106
Query: 594 GPDGGSLAKLTVDYHTQGDASPSEEEIKAGQAQGDGLFKAVEGYVLAHPDY 442
GPDGGS+AKLTV Y T+GDA+PSEEEIK G+A+GDGLFKA+EGYVLA+PDY
Sbjct: 107 GPDGGSIAKLTVKYFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>sp|Q43560|PR1_MEDSA Class-10 pathogenesis-related protein 1 (MSPR10-1)
gi|7442207|pir||T09659 pathogenesis-related protein
class 10 - alfalfa gi|1419683|emb|CAA67375.1| PR protein
from class 10 [Medicago sativa]
Length = 157
Score = 164 bits (415), Expect = 1e-39
Identities = 77/111 (69%), Positives = 95/111 (85%)
Frame = -3
Query: 774 NGSAGTIKKITFEEDGQTKDLLHQIESIDEAHFGYSYSIIGGSDLPDPVEKISFEAQLVA 595
NG AGTIKK+TF E G+TK LH+++ +D+ +F Y+YSI+GG LPD VEKISFE++L A
Sbjct: 47 NGGAGTIKKLTFVEGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSA 106
Query: 594 GPDGGSLAKLTVDYHTQGDASPSEEEIKAGQAQGDGLFKAVEGYVLAHPDY 442
GPDGGS AKLTV Y T+GDA+PSEEEIK G+A+GDGLFKA+EGYVLA+PDY
Sbjct: 107 GPDGGSTAKLTVKYFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>pir||T09526 stress response gene 1 - alfalfa gi|1136333|gb|AAB58315.1| Srg1
[Medicago sativa]
Length = 157
Score = 162 bits (410), Expect = 5e-39
Identities = 77/111 (69%), Positives = 94/111 (84%)
Frame = -3
Query: 774 NGSAGTIKKITFEEDGQTKDLLHQIESIDEAHFGYSYSIIGGSDLPDPVEKISFEAQLVA 595
NG AGTIKK TF E G+TK LH+++ +D+ +F Y+YSI+GG LPD VEKISFE++L A
Sbjct: 47 NGGAGTIKKPTFVEGGETKYDLHKVDLVDDVNFAYNYSIVGGGGLPDTVEKISFESKLSA 106
Query: 594 GPDGGSLAKLTVDYHTQGDASPSEEEIKAGQAQGDGLFKAVEGYVLAHPDY 442
GPDGGS AKLTV Y T+GDA+PSEEEIK G+A+GDGLFKA+EGYVLA+PDY
Sbjct: 107 GPDGGSTAKLTVKYFTKGDAAPSEEEIKGGKARGDGLFKALEGYVLANPDY 157
>sp|P14710|DRR3_PEA Disease resistance response protein Pi49 (PR10)
gi|7442191|pir||T06527 pathogenesis-related protein 10 -
garden pea gi|436313|emb|CAA31760.1| disease resistance
response protein [Pisum sativum]
gi|967270|gb|AAA90954.1| PR10 gi|226758|prf||1604467A
disease response resistance gene
Length = 158
Score = 161 bits (408), Expect = 9e-39
Identities = 74/112 (66%), Positives = 95/112 (84%)
Frame = -3
Query: 774 NGSAGTIKKITFEEDGQTKDLLHQIESIDEAHFGYSYSIIGGSDLPDPVEKISFEAQLVA 595
NG AGTIKK+TF EDG+TK +LH++E +D A+ Y+YSI+GG PD VEKISFEA+L A
Sbjct: 47 NGGAGTIKKLTFVEDGETKHVLHKVELVDVANLAYNYSIVGGVGFPDTVEKISFEAKLSA 106
Query: 594 GPDGGSLAKLTVDYHTQGDASPSEEEIKAGQAQGDGLFKAVEGYVLAHPDYH 439
GP+GGS+AKL+V Y T+GDA+PSEE++K +A+GDGLFKA+EGY LAHPDY+
Sbjct: 107 GPNGGSIAKLSVKYFTKGDAAPSEEQLKTDKAKGDGLFKALEGYCLAHPDYN 158
>sp|P26987|SAM2_SOYBN Stress-induced protein SAM22 gi|99918|pir||S20518 hypothetical
protein - soybean gi|18744|emb|CAA42646.1| ORF [Glycine
max]
Length = 158
Score = 161 bits (408), Expect = 9e-39
Identities = 73/112 (65%), Positives = 95/112 (84%)
Frame = -3
Query: 774 NGSAGTIKKITFEEDGQTKDLLHQIESIDEAHFGYSYSIIGGSDLPDPVEKISFEAQLVA 595
NG GTIKKITF EDG+TK +LH+IESIDEA+ GYSYS++GG+ LPD EKI+F+++LVA
Sbjct: 47 NGGPGTIKKITFLEDGETKFVLHKIESIDEANLGYSYSVVGGAALPDTAEKITFDSKLVA 106
Query: 594 GPDGGSLAKLTVDYHTQGDASPSEEEIKAGQAQGDGLFKAVEGYVLAHPDYH 439
GP+GGS KLTV Y T+GDA P+++E+K G+A+ D LFKA+E Y+LAHPDY+
Sbjct: 107 GPNGGSAGKLTVKYETKGDAEPNQDELKTGKAKADALFKAIEAYLLAHPDYN 158
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 756,061,672
Number of Sequences: 1393205
Number of extensions: 18766640
Number of successful extensions: 52491
Number of sequences better than 10.0: 238
Number of HSP's better than 10.0 without gapping: 48914
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 52322
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38375267554
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)