Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
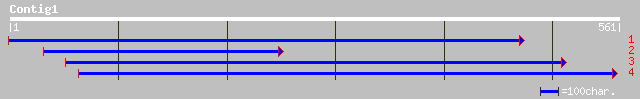
Query= KMC004262A_C01 KMC004262A_c01
(561 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_564894.1| F-box protein family; protein id: At1g67340.1, ... 128 5e-29
ref|NP_199856.1| putative protein; protein id: At5g50450.1, supp... 107 8e-23
dbj|BAB86182.1| OJ1485_B09.11 [Oryza sativa (japonica cultivar-g... 94 1e-18
gb|AAH47974.1| Similar to programmed cell death 2 [Xenopus laevis] 50 2e-05
ref|NP_496323.1| SET-domain transcriptional regulator like (48.5... 48 9e-05
>ref|NP_564894.1| F-box protein family; protein id: At1g67340.1, supported by cDNA:
gi_16209725, supported by cDNA: gi_20260405 [Arabidopsis
thaliana] gi|25373198|pir||H96696 protein F1N21.16
[imported] - Arabidopsis thaliana
gi|9828618|gb|AAG00241.1|AC002130_6 F1N21.16
[Arabidopsis thaliana] gi|16209726|gb|AAL14418.1|
At1g67340/F1N21_16 [Arabidopsis thaliana]
gi|20260406|gb|AAM13101.1| unknown protein [Arabidopsis
thaliana] gi|23197876|gb|AAN15465.1| unknown protein
[Arabidopsis thaliana]
Length = 379
Score = 128 bits (321), Expect = 5e-29
Identities = 59/85 (69%), Positives = 68/85 (79%), Gaps = 1/85 (1%)
Frame = -3
Query: 559 NRFMTEWFAAKD-DVPGPGLSLCSHMGCGRPETRNREFRRCSVCGTVNYCSRACQALDWK 383
NRF+ +WFA + D PG GL LCSH GCGRPETR EFRRCSVCG VNYCSRACQALDWK
Sbjct: 280 NRFLADWFAVRGGDCPGDGLRLCSHAGCGRPETRKHEFRRCSVCGVVNYCSRACQALDWK 339
Query: 382 LRHKVECAAPMERLLDDDGDDAGEG 308
LRHK++C AP++R L++ D GEG
Sbjct: 340 LRHKMDC-APVQRWLEE--GDGGEG 361
>ref|NP_199856.1| putative protein; protein id: At5g50450.1, supported by cDNA:
gi_20466553 [Arabidopsis thaliana]
gi|9758927|dbj|BAB09464.1|
gb|AAB95234.1~gene_id:MXI22.17~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|20466554|gb|AAM20594.1| putative protein [Arabidopsis
thaliana] gi|23198134|gb|AAN15594.1| putative protein
[Arabidopsis thaliana]
Length = 336
Score = 107 bits (268), Expect = 8e-23
Identities = 53/98 (54%), Positives = 62/98 (63%), Gaps = 7/98 (7%)
Frame = -3
Query: 559 NRFMTEWFAAKDDVPGPGLSLCSHMGCGRPETRNREFRRCSVCGTVNYCSRACQALDWKL 380
NRF+ EWF++ GL +CSH GCGRPETR EFRRCSVCG VNYCSR CQALDW+
Sbjct: 240 NRFLKEWFSSGRVDLAEGLRMCSHGGCGRPETRAHEFRRCSVCGKVNYCSRGCQALDWRA 299
Query: 379 RHKVEC-------AAPMERLLDDDGDDAGEGVGDGEVE 287
+HKVEC AA E + DDG+ + D E
Sbjct: 300 KHKVECTPLDLWVAAAAE--IGDDGEAVAVEIDDNHGE 335
>dbj|BAB86182.1| OJ1485_B09.11 [Oryza sativa (japonica cultivar-group)]
Length = 388
Score = 93.6 bits (231), Expect = 1e-18
Identities = 47/93 (50%), Positives = 54/93 (57%), Gaps = 19/93 (20%)
Frame = -3
Query: 559 NRFMTEWFAAK-------------------DDVPGPGLSLCSHMGCGRPETRNREFRRCS 437
N FM +W+A++ D G L LCSH+ CGR ETR EFRRCS
Sbjct: 290 NLFMADWWASRGVQATAKKPGLEAPAAATGDSDGGGELRLCSHVRCGRRETRRHEFRRCS 349
Query: 436 VCGTVNYCSRACQALDWKLRHKVECAAPMERLL 338
VCG NYCSRACQALDWK HK +C PM+R L
Sbjct: 350 VCGAANYCSRACQALDWKRAHKAQC-VPMDRWL 381
>gb|AAH47974.1| Similar to programmed cell death 2 [Xenopus laevis]
Length = 361
Score = 49.7 bits (117), Expect = 2e-05
Identities = 24/50 (48%), Positives = 29/50 (58%)
Frame = -3
Query: 508 GLSLCSHMGCGRPETRNREFRRCSVCGTVNYCSRACQALDWKLRHKVECA 359
GL LC GC P+T CS C VNYCS+ Q +DWKL+HK C+
Sbjct: 132 GLRLCRVCGCLGPKT-------CSKCHKVNYCSKDHQLMDWKLQHKKVCS 174
>ref|NP_496323.1| SET-domain transcriptional regulator like (48.5 kD) [Caenorhabditis
elegans] gi|3123316|sp|Q09415|YRM4_CAEEL Hypothetical
protein R06F6.4 in chromosome II gi|7506368|pir||T23984
hypothetical protein R06F6.4 - Caenorhabditis elegans
gi|3878909|emb|CAA86783.1| Hypothetical protein R06F6.4
[Caenorhabditis elegans]
Length = 429
Score = 47.8 bits (112), Expect = 9e-05
Identities = 21/42 (50%), Positives = 26/42 (61%)
Frame = -3
Query: 481 CGRPETRNREFRRCSVCGTVNYCSRACQALDWKLRHKVECAA 356
C + T E ++CS C + YCS+ CQ DWKL HKVEC A
Sbjct: 26 CNQCLTSMAELKKCSACRRLAYCSQECQRADWKL-HKVECKA 66
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,123,798
Number of Sequences: 1393205
Number of extensions: 14381095
Number of successful extensions: 95841
Number of sequences better than 10.0: 786
Number of HSP's better than 10.0 without gapping: 66646
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 89359
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)