Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004245A_C01 KMC004245A_c01
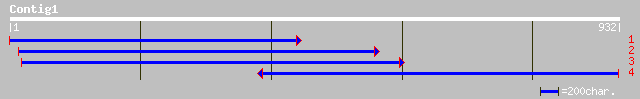
(932 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563826.1| expressed protein; protein id: At1g08770.1, sup... 170 3e-41
gb|AAM66031.1| unknown [Arabidopsis thaliana] 167 2e-40
gb|AAM66945.1| unknown [Arabidopsis thaliana] 145 1e-33
ref|NP_566461.1| expressed protein; protein id: At3g13720.1, sup... 144 2e-33
ref|NP_564679.1| expressed protein; protein id: At1g55190.1, sup... 138 1e-31
>ref|NP_563826.1| expressed protein; protein id: At1g08770.1, supported by cDNA:
7188. [Arabidopsis thaliana] gi|7485990|pir||T00733
hypothetical protein F22O13.28 - Arabidopsis thaliana
gi|9802565|gb|AAF99767.1|AC003981_17 F22O13.26
[Arabidopsis thaliana] gi|18072829|emb|CAC80647.1|
prenylated Rab receptor 4 [Arabidopsis thaliana]
Length = 209
Score = 170 bits (430), Expect = 3e-41
Identities = 92/187 (49%), Positives = 124/187 (66%), Gaps = 6/187 (3%)
Frame = -2
Query: 928 GTVSAATTTTSQPPPPSTSNLIATPRSWREFLDISALSRPYSYDDAMIRLRRNLSYFRFN 749
GT+SA T+Q ++I T R WRE LD+SALS P YD+AM L+ N+SYFR N
Sbjct: 30 GTLSARAKQTTQ-------SMITTLRPWREILDLSALSLPRGYDEAMAHLKHNISYFRGN 82
Query: 748 YAAVSLLIIFLTLLWHPVSMIVFLILLVAWFFLYFSRDG--PLVVLNRTVDDRTVLCVLG 575
YA L I+FL L++HP+SMI F+++ + W LYFSRD +V+ + VDD+ VL +L
Sbjct: 83 YALAVLAIVFLGLIYHPMSMIAFIVVFIGWILLYFSRDANDSIVISGKEVDDKIVLVLLS 142
Query: 574 LVTVVGLVWTHVGLNVLVALIVCVVVVGLHAAFRVIEDLFHDEES----GLLSVVGSQPL 407
LVTV+ LV+T VG NVLV+LI+ +++VG H AFR +DLF DEES GL+S
Sbjct: 143 LVTVLALVYTDVGENVLVSLIIGLLIVGAHGAFRNTDDLFLDEESARRGGLVSAGSGNRP 202
Query: 406 RTNYTPI 386
++YTPI
Sbjct: 203 PSSYTPI 209
>gb|AAM66031.1| unknown [Arabidopsis thaliana]
Length = 209
Score = 167 bits (424), Expect = 2e-40
Identities = 91/187 (48%), Positives = 123/187 (65%), Gaps = 6/187 (3%)
Frame = -2
Query: 928 GTVSAATTTTSQPPPPSTSNLIATPRSWREFLDISALSRPYSYDDAMIRLRRNLSYFRFN 749
GT+SA T+Q ++I T R WRE LD+SALS P YD+AM L+ N+SYFR N
Sbjct: 30 GTLSARAKQTTQ-------SMITTLRPWREILDLSALSLPRGYDEAMAHLKHNISYFRGN 82
Query: 748 YAAVSLLIIFLTLLWHPVSMIVFLILLVAWFFLYFSRDG--PLVVLNRTVDDRTVLCVLG 575
YA L I+FL L++HP+SMI F+++ + W LYFSRD +V+ + VDD+ VL +L
Sbjct: 83 YALAVLAIVFLGLIYHPMSMIAFIVVFIGWILLYFSRDANDSIVISGKEVDDKIVLVLLS 142
Query: 574 LVTVVGLVWTHVGLNVLVALIVCVVVVGLHAAFRVIEDLFHDEES----GLLSVVGSQPL 407
LVTV+ LV+T VG NVLV+LI+ +++VG H AFR +DLF DEES L+S
Sbjct: 143 LVTVLALVYTDVGENVLVSLIIGLLIVGAHGAFRNTDDLFLDEESARRGSLVSAGSGNRP 202
Query: 406 RTNYTPI 386
++YTPI
Sbjct: 203 PSSYTPI 209
>gb|AAM66945.1| unknown [Arabidopsis thaliana]
Length = 188
Score = 145 bits (365), Expect = 1e-33
Identities = 69/149 (46%), Positives = 97/149 (64%)
Frame = -2
Query: 865 IATPRSWREFLDISALSRPYSYDDAMIRLRRNLSYFRFNYAAVSLLIIFLTLLWHPVSMI 686
+AT R+WR D ++ P+ DA R++ NL+YFR NYA V L++IF +L+WHP S+I
Sbjct: 33 LATRRAWRVMFDFHSMGLPHGVSDAFTRIKTNLAYFRMNYAIVVLIVIFFSLIWHPTSLI 92
Query: 685 VFLILLVAWFFLYFSRDGPLVVLNRTVDDRTVLCVLGLVTVVGLVWTHVGLNVLVALIVC 506
VF +L+V W FLYF RD P+ + +DDRTVL VL ++TVV L+ T+ N++ AL+
Sbjct: 93 VFTVLVVVWIFLYFLRDEPIKLFRFQIDDRTVLIVLSVLTVVLLLLTNATFNIVGALVTG 152
Query: 505 VVVVGLHAAFRVIEDLFHDEESGLLSVVG 419
V+V +HA R EDLF DEE+ G
Sbjct: 153 AVLVLIHAVVRKTEDLFLDEEAATTETSG 181
>ref|NP_566461.1| expressed protein; protein id: At3g13720.1, supported by cDNA:
8544. [Arabidopsis thaliana] gi|9294018|dbj|BAB01921.1|
gb|AAC14054.1~gene_id:MMM17.14~similar to unknown
protein [Arabidopsis thaliana]
gi|28393547|gb|AAO42194.1| unknown protein [Arabidopsis
thaliana] gi|28827282|gb|AAO50485.1| unknown protein
[Arabidopsis thaliana]
Length = 188
Score = 144 bits (362), Expect = 2e-33
Identities = 68/149 (45%), Positives = 97/149 (64%)
Frame = -2
Query: 865 IATPRSWREFLDISALSRPYSYDDAMIRLRRNLSYFRFNYAAVSLLIIFLTLLWHPVSMI 686
+AT R+WR D ++ P+ DA R++ NL+YFR NYA V L++IF +L+WHP S+I
Sbjct: 33 LATRRAWRVMFDFHSMGLPHGVSDAFTRIKTNLAYFRMNYAIVVLIVIFFSLIWHPTSLI 92
Query: 685 VFLILLVAWFFLYFSRDGPLVVLNRTVDDRTVLCVLGLVTVVGLVWTHVGLNVLVALIVC 506
VF +L+V W FLYF RD P+ + +DDRTVL VL ++TVV L+ T+ N++ AL+
Sbjct: 93 VFTVLVVVWIFLYFLRDEPIKLFRFQIDDRTVLIVLSVLTVVLLLLTNATFNIVGALVTG 152
Query: 505 VVVVGLHAAFRVIEDLFHDEESGLLSVVG 419
V+V +H+ R EDLF DEE+ G
Sbjct: 153 AVLVLIHSVVRKTEDLFLDEEAATTETSG 181
>ref|NP_564679.1| expressed protein; protein id: At1g55190.1, supported by cDNA:
21940. [Arabidopsis thaliana] gi|25347725|pir||F96593
hypothetical protein F7A10.20 [imported] - Arabidopsis
thaliana gi|12323168|gb|AAG51564.1|AC027034_10
hypothetical protein; 89971-89402 [Arabidopsis thaliana]
gi|21554296|gb|AAM63371.1| unknown [Arabidopsis
thaliana] gi|28393384|gb|AAO42116.1| unknown protein
[Arabidopsis thaliana] gi|28827574|gb|AAO50631.1|
unknown protein [Arabidopsis thaliana]
Length = 189
Score = 138 bits (348), Expect = 1e-31
Identities = 74/157 (47%), Positives = 101/157 (64%), Gaps = 7/157 (4%)
Frame = -2
Query: 865 IATPRSWREFLDISALSRPYSYDDAMIRLRRNLSYFRFNYAAVSLLIIFLTLLWHPVSMI 686
+AT R W+ D +++ P+ + DA+ R++ NL YFR NYA L I+FL+LL+HP S+I
Sbjct: 33 LATRRPWKSMFDFESMTLPHGFFDAISRIKTNLGYFRANYAIGVLFILFLSLLYHPTSLI 92
Query: 685 VFLILLVAWFFLYFSRDGPLVVLNRTVDDRTVLCVLGLVTVVGLVWTHVGLNVLVALIVC 506
V IL+V W FLYF RD PLVV +DDRTVL L ++TVV L+ TH N+L +L+
Sbjct: 93 VLSILVVFWIFLYFLRDEPLVVFGYQIDDRTVLIGLSVLTVVMLLLTHATSNILGSLLTA 152
Query: 505 VVVVGLHAAFRVIEDLFHDEE-------SGLLSVVGS 416
V+V +HAA R ++LF DEE SGL+S S
Sbjct: 153 AVLVLIHAAVRRSDNLFLDEEAAAVTEASGLMSYPSS 189
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 941,213,653
Number of Sequences: 1393205
Number of extensions: 27220571
Number of successful extensions: 399010
Number of sequences better than 10.0: 7172
Number of HSP's better than 10.0 without gapping: 138767
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 281521
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 51859780984
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)