Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004237A_C01 KMC004237A_c01
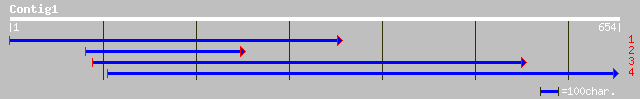
(652 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB86205.1| B1147A04.22 [Oryza sativa (japonica cultivar-gro... 78 1e-13
ref|XP_162816.2| similar to cyclin-dependent kinase 4 [Rattus no... 53 3e-06
ref|NP_062602.1| proline-rich protein MP5; proline-rich protein ... 52 7e-06
pir||JH0481 basic proline-rich protein MnP4 - crab-eating macaqu... 50 3e-05
ref|NP_196852.1| putative protein; protein id: At5g13480.1 [Arab... 50 3e-05
>dbj|BAB86205.1| B1147A04.22 [Oryza sativa (japonica cultivar-group)]
Length = 593
Score = 77.8 bits (190), Expect = 1e-13
Identities = 38/73 (52%), Positives = 46/73 (62%), Gaps = 4/73 (5%)
Frame = -3
Query: 590 QGAMNQMGNPMPQGPFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSNT----NYPQGAS 423
QG+ NQM MPQ +GMNQ H GPPS PP+GGFPN + N+QG S+ N+P G
Sbjct: 505 QGSSNQMMPQMPQH-LIGMNQTHQGPPSNMPPMGGFPNGMGNIQGGSSASGMQNFPMGGM 563
Query: 422 FNRPQGGQMPMMQ 384
+NRPQG P Q
Sbjct: 564 YNRPQGKMPPQAQ 576
>ref|XP_162816.2| similar to cyclin-dependent kinase 4 [Rattus norvegicus] [Mus
musculus]
Length = 247
Score = 53.1 bits (126), Expect = 3e-06
Identities = 46/115 (40%), Positives = 47/115 (40%), Gaps = 14/115 (12%)
Frame = -3
Query: 623 PMPMPGLMGMQQGAMNQMGN---PMPQG------PFVGMNQMHSGPPSGGPPLGGFPNNL 471
P P P G Q Q GN P PQG P G Q PP GGPP G P
Sbjct: 46 PPPGPPPQGGPQQRPPQPGNQQGPPPQGGHQQRPPQPGNQQ--GPPPQGGPPPQGGPQRP 103
Query: 470 P---NMQGPSNTNYPQGASFNRPQGGQMPMMQG--YNPYQSGNQPGMPPNAQPGG 321
P N QGP PQG RP Q P QG P + GNQ G PP P G
Sbjct: 104 PQPGNQQGPPPQGGPQGPP--RPGNQQGPPPQGGPQGPPRPGNQQGPPPQGGPQG 156
Score = 48.9 bits (115), Expect = 6e-05
Identities = 43/104 (41%), Positives = 44/104 (41%), Gaps = 7/104 (6%)
Frame = -3
Query: 611 PGLMGMQQGAMNQMGNPMPQG-----PFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSN 447
P G QQG Q G P PQG P G Q GPP G P G P N QGP
Sbjct: 79 PPQPGNQQGPPPQ-GGPPPQGGPQRPPQPGNQQ---GPPPQGGPQG--PPRPGNQQGPPP 132
Query: 446 TNYPQGASFNRPQGGQMPMMQG--YNPYQSGNQPGMPPNAQPGG 321
PQG RP Q P QG P + GNQ G PP P G
Sbjct: 133 QGGPQGPP--RPGNQQGPPPQGGPQGPPRPGNQQGPPPQGGPQG 174
Score = 48.5 bits (114), Expect = 8e-05
Identities = 36/97 (37%), Positives = 36/97 (37%), Gaps = 16/97 (16%)
Frame = -3
Query: 563 PMPQG-----PFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSNTNYPQGASFNR----- 414
P PQG P G Q GPP GPP G P P G PQG R
Sbjct: 27 PPPQGGPQSPPRPGTQQ---GPPPPGPPPQGGPQQRPPQPGNQQGPPPQGGHQQRPPQPG 83
Query: 413 ------PQGGQMPMMQGYNPYQSGNQPGMPPNAQPGG 321
PQGG P P Q GNQ G PP P G
Sbjct: 84 NQQGPPPQGGPPPQGGPQRPPQPGNQQGPPPQGGPQG 120
>ref|NP_062602.1| proline-rich protein MP5; proline-rich protein PRP MP5 [Mus
musculus] gi|110849|pir||S22373 proline-rich protein -
mouse gi|53797|emb|CAA44734.1| proline-rich protein [Mus
musculus] gi|53799|emb|CAA44733.1| proline-rich protein
[Mus musculus]
Length = 260
Score = 52.0 bits (123), Expect = 7e-06
Identities = 44/129 (34%), Positives = 48/129 (37%), Gaps = 13/129 (10%)
Frame = -3
Query: 650 GISSVXTSHPMPMPGLMGMQQGAMNQMGNPMPQGPFVGMNQ------MHSGPPSGGPPLG 489
G S+ S P P P G P P+GP G Q GPP GPP
Sbjct: 33 GHSTTVVSDPSPTPQPRPKHSG-------PPPKGPRPGSTQGPPRVPNQQGPPPPGPPPQ 85
Query: 488 GFPNNLP----NMQGPSNTNYPQGASFNRPQGGQMPMMQG---YNPYQSGNQPGMPPNAQ 330
G P N QGP PQG RP Q+P Q P Q GNQ G PP
Sbjct: 86 GSSQQRPPQPGNQQGPPPQGGPQGPP--RPGNQQVPPPQEGSQQRPPQPGNQQGPPPQGP 143
Query: 329 PGGSHNQ*P 303
P + Q P
Sbjct: 144 PRPGNQQGP 152
Score = 48.5 bits (114), Expect = 8e-05
Identities = 42/113 (37%), Positives = 44/113 (38%), Gaps = 5/113 (4%)
Frame = -3
Query: 623 PMPMPGLMGMQQGAMNQMGNPMPQGPFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSNT 444
P P G QQG Q G PQGP NQ PP GGP P N QGP
Sbjct: 139 PPQGPPRPGNQQGPPPQGG---PQGPSRPGNQQ-GPPPQGGPQQR--PPQPGNQQGPPPQ 192
Query: 443 NYPQGASFNRPQGGQMPMMQGYNPYQSGNQPGMPPNAQP-----GGSHNQ*PQ 300
PQG Q G P P + GNQ G PP P G+H PQ
Sbjct: 193 GGPQGGPQPGNQQGPPPQGGPQGPPRPGNQQGPPPQGGPQRPPQPGNHQGPPQ 245
Score = 48.5 bits (114), Expect = 8e-05
Identities = 46/128 (35%), Positives = 47/128 (35%), Gaps = 27/128 (21%)
Frame = -3
Query: 623 PMPMPGLMGMQQGAMNQMGN---PMPQG-----PFVGMNQM----------------HSG 516
P P P G Q Q GN P PQG P G Q+ G
Sbjct: 78 PPPGPPPQGSSQQRPPQPGNQQGPPPQGGPQGPPRPGNQQVPPPQEGSQQRPPQPGNQQG 137
Query: 515 PPSGGPPLGGFPNNLPNMQGPSNTNYPQGASFNRPQGGQMPMMQG---YNPYQSGNQPGM 345
PP GPP G N QGP PQG S RP Q P QG P Q GNQ G
Sbjct: 138 PPPQGPPRPG------NQQGPPPQGGPQGPS--RPGNQQGPPPQGGPQQRPPQPGNQQGP 189
Query: 344 PPNAQPGG 321
PP P G
Sbjct: 190 PPQGGPQG 197
Score = 45.4 bits (106), Expect = 6e-04
Identities = 34/104 (32%), Positives = 38/104 (35%), Gaps = 5/104 (4%)
Frame = -3
Query: 617 PMPGLMGMQQGAMNQMGNPMPQGPFVGMNQMHSGPPSG--GPPLGGFPNNLPNMQGPSNT 444
P PG NQ G P P P G +Q P GPP G P P P N
Sbjct: 60 PRPGSTQGPPRVPNQQGPPPPGPPPQGSSQQRPPQPGNQQGPPPQGGPQGPPR---PGNQ 116
Query: 443 NYP---QGASFNRPQGGQMPMMQGYNPYQSGNQPGMPPNAQPGG 321
P +G+ PQ G P + GNQ G PP P G
Sbjct: 117 QVPPPQEGSQQRPPQPGNQQGPPPQGPPRPGNQQGPPPQGGPQG 160
>pir||JH0481 basic proline-rich protein MnP4 - crab-eating macaque
gi|342283|gb|AAA36904.1| proline-rich protein
gi|342285|gb|AAA36905.1| proline-rich protein
Length = 188
Score = 50.1 bits (118), Expect = 3e-05
Identities = 41/120 (34%), Positives = 46/120 (38%), Gaps = 9/120 (7%)
Frame = -3
Query: 641 SVXTSHPMPMPGLMGMQQGAMNQMGNPMPQGPFVGMNQMHSGPPSGGPPLGGFPNNLPNM 462
S+ + H P QQG G P P G G Q P G PP G P P
Sbjct: 30 SLISEHQQGQP-----QQGGNRPQGPPSPPGNAQGPPQQGGKKPQGPPP-PGKPQGPPKQ 83
Query: 461 -----QGPSNTNYPQG---ASFNRPQGGQMPMMQGYNPYQSGNQP-GMPPNAQPGGSHNQ 309
QGP PQG N+PQG P P Q GN+P G PP +P G Q
Sbjct: 84 GGKKPQGPPPPGKPQGPPQQGGNKPQGPPPPGKPQGPPQQGGNKPQGPPPPGKPQGPPQQ 143
Score = 43.9 bits (102), Expect = 0.002
Identities = 35/101 (34%), Positives = 38/101 (36%), Gaps = 8/101 (7%)
Frame = -3
Query: 593 QQGAMNQMGNP---MPQGPFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSNTNYPQG-- 429
QQG G P PQGP + GPP G P G QGP PQG
Sbjct: 62 QQGGKKPQGPPPPGKPQGPPKQGGKKPQGPPPPGKPQGPPQQGGNKPQGPPPPGKPQGPP 121
Query: 428 -ASFNRPQGGQMPMMQGYNPYQSGN--QPGMPPNAQPGGSH 315
N+PQG P P Q GN Q PP +P H
Sbjct: 122 QQGGNKPQGPPPPGKPQGPPQQGGNAQQAQPPPAGRPRDHH 162
>ref|NP_196852.1| putative protein; protein id: At5g13480.1 [Arabidopsis thaliana]
gi|9955540|emb|CAC05425.1| putative protein [Arabidopsis
thaliana]
Length = 679
Score = 49.7 bits (117), Expect = 3e-05
Identities = 39/105 (37%), Positives = 44/105 (41%), Gaps = 6/105 (5%)
Frame = -3
Query: 614 MPGLMGMQQGAMNQMGNP----MPQGPFVGMNQMHSGPPSGGPPLGGFPNNLPNMQGPSN 447
MPG MGMQ G QM P G F G P SGGP +
Sbjct: 599 MPGSMGMQGGMNPQMSQSHFMGAPSGVFQGQ------PNSGGPQM--------------- 637
Query: 446 TNYPQG-ASFNRPQGGQMPMMQGYN-PYQSGNQPGMPPNAQPGGS 318
YPQG FNRPQ M+ GYN P+Q QP +PP P +
Sbjct: 638 --YPQGRGGFNRPQ-----MIPGYNNPFQQQQQPPLPPGPPPNNN 675
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 580,141,530
Number of Sequences: 1393205
Number of extensions: 14255133
Number of successful extensions: 48310
Number of sequences better than 10.0: 1046
Number of HSP's better than 10.0 without gapping: 38696
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 45605
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)