Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
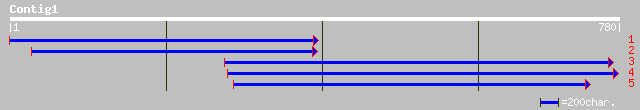
Query= KMC004230A_C01 KMC004230A_c01
(774 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM33190.2| similar to Dictyostelium discoideum (Slime mold).... 41 0.022
gb|AAO51288.1| similar to Dictyostelium discoideum (Slime mold).... 40 0.029
gb|AAB36702.1| putative transcription factor [Dictyostelium disc... 40 0.029
ref|NP_705527.1| hypothetical protein [Plasmodium falciparum 3D7... 40 0.037
gb|AAD30963.2| SNF1/AMP-activated kinase [Dictyostelium discoideum] 40 0.037
>gb|AAM33190.2| similar to Dictyostelium discoideum (Slime mold). Histidine kinase
DhkE
Length = 619
Score = 40.8 bits (94), Expect = 0.022
Identities = 35/127 (27%), Positives = 49/127 (38%)
Frame = +1
Query: 394 ITENQVITSVHPHPHLMADERQKQQDKQQVKGLTHALNINQIAS*IKKQHIADKG*SS*M 573
I +NQ P ++Q+QQ +QQ + Q IK + +K +
Sbjct: 59 INQNQPEKRSLPTKDEKQQQQQQQQQQQQQQQQQQQQQQQQQEKQIKNKEKDEKDEKEEI 118
Query: 574 SICSLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINNCTSKGGT 753
I + N S NNNNNNN NN N + +NNNN NN +
Sbjct: 119 KIEIIDNNSNNNNNNNNNNNNNNNNNNNNNNNNNNN---------NNNNNNNNNNNNEDK 169
Query: 754 VTERKQK 774
+ E K K
Sbjct: 170 IKENKVK 176
>gb|AAO51288.1| similar to Dictyostelium discoideum (Slime mold). Putative
transcription factor
Length = 844
Score = 40.4 bits (93), Expect = 0.029
Identities = 23/61 (37%), Positives = 32/61 (51%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINNCTSKGGTVTE 762
S NGS NNNN+NN NN N++ ++S+ NNNN NN TS+ + +
Sbjct: 604 SNNNGSYNNNNSNNN---NNNNRIEPIVNNNINNFNSINNNNSNNNNNNNSTSQNAPLLQ 660
Query: 763 R 765
R
Sbjct: 661 R 661
Score = 33.1 bits (74), Expect = 4.6
Identities = 18/50 (36%), Positives = 25/50 (50%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
+L+N NNNNNNN NN + +++ D+ NNNN NN
Sbjct: 58 NLENNIDNNNNNNNNNNNNNNNNNNNNSKSIINLIDNNNNNNKNNNNNNN 107
>gb|AAB36702.1| putative transcription factor [Dictyostelium discoideum]
Length = 872
Score = 40.4 bits (93), Expect = 0.029
Identities = 23/61 (37%), Positives = 32/61 (51%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINNCTSKGGTVTE 762
S NGS NNNN+NN NN N++ ++S+ NNNN NN TS+ + +
Sbjct: 632 SNNNGSYNNNNSNNN---NNNNRIEPIVNNNINNFNSINNNNSNNNNNNNSTSQNAPLLQ 688
Query: 763 R 765
R
Sbjct: 689 R 689
Score = 33.1 bits (74), Expect = 4.6
Identities = 18/50 (36%), Positives = 25/50 (50%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
+L+N NNNNNNN NN + +++ D+ NNNN NN
Sbjct: 86 NLENNIDNNNNNNNNNNNNNNNNNNNNSKSIINLIDNNNNNNKNNNNNNN 135
>ref|NP_705527.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23615772|emb|CAD52764.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 3265
Score = 40.0 bits (92), Expect = 0.037
Identities = 23/66 (34%), Positives = 28/66 (41%)
Frame = +1
Query: 577 ICSLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINNCTSKGGTV 756
I + N S NNNNNNN NN +NNNN NNC GG V
Sbjct: 1565 ISAHHNNSNNNNNNNNNNNNNNNN--------------------NNNNNNNNCNKDGGVV 1604
Query: 757 TERKQK 774
R+++
Sbjct: 1605 RTRRKE 1610
Score = 32.7 bits (73), Expect = 6.0
Identities = 18/47 (38%), Positives = 24/47 (50%)
Frame = +1
Query: 592 NGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
N + NNNNNNN NN I + N + ++ +NNNN NN
Sbjct: 2841 NNNNNNNNNNNNNNNNNNNINNNNSNNNNNNNNNNNNNNNNNNNNNN 2887
>gb|AAD30963.2| SNF1/AMP-activated kinase [Dictyostelium discoideum]
Length = 718
Score = 40.0 bits (92), Expect = 0.037
Identities = 22/57 (38%), Positives = 30/57 (52%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINNCTSKGGT 753
++ N +INNNNNNN NN I N + ++ +NNNN NN + GGT
Sbjct: 429 NINNNNINNNNNNNNNNINNNNIINNNNNNNNNNNNNNNNNNNNNNNNNNSSISGGT 485
Score = 35.8 bits (81), Expect = 0.70
Identities = 18/47 (38%), Positives = 23/47 (48%)
Frame = +1
Query: 592 NGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
N NNNNNNN NN N + DS+ L+N N++NN
Sbjct: 520 NNDNNNNNNNNNNNNNNNNNNNNNNNNNNNCIDSVNNSLNNENDVNN 566
Score = 33.9 bits (76), Expect = 2.7
Identities = 19/50 (38%), Positives = 23/50 (46%)
Frame = +1
Query: 583 SLQNGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
S+ N + NNNNNNN NN T + + NNNNINN
Sbjct: 395 SISNNNNNNNNNNNNTTNNNNNTT------------NNNNSIINNNNINN 432
Score = 33.5 bits (75), Expect = 3.5
Identities = 17/47 (36%), Positives = 24/47 (50%)
Frame = +1
Query: 592 NGSINNNNNNNTPMGNNGTITVGAT*KNLHMWDSLRLPLDNNNNINN 732
N + NNNNNNN NN + + +L+ + + NNNN NN
Sbjct: 530 NNNNNNNNNNNNNNNNNNNNCIDSVNNSLNNENDVNNSNINNNNNNN 576
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 668,634,994
Number of Sequences: 1393205
Number of extensions: 15352984
Number of successful extensions: 123247
Number of sequences better than 10.0: 379
Number of HSP's better than 10.0 without gapping: 42655
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 71030
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38095156112
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)