Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004216A_C01 KMC004216A_c01
(973 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
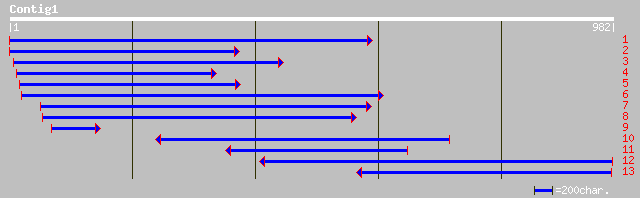
pir||T11744 dehydrin - kidney bean gi|1326161|gb|AAB00554.1| deh... 245 9e-64
emb|CAC35772.3| dhn1 [Populus euramericana] 203 4e-51
pir||S50766 dehydrin-related protein - garden pea gi|18375|emb|C... 201 1e-50
gb|AAN78125.1| dehydrin [Citrus x paradisi] 195 1e-48
gb|AAF01465.2|AF190474_1 bdn1 [Boea crassifolia] 189 4e-47
>pir||T11744 dehydrin - kidney bean gi|1326161|gb|AAB00554.1| dehydrin
Length = 202
Score = 245 bits (625), Expect = 9e-64
Identities = 149/217 (68%), Positives = 168/217 (76%), Gaps = 12/217 (5%)
Frame = -3
Query: 869 KENQNKYESG-TTEVETQDRGVFDFLGKKKEEEKPQEEVIVTELEKVSVSEEPEKKKEEV 693
+E QNKYE+ ++EVE QDRGVFDFLGKKKEEEKPQEEVIVTE EK++VSEE KKEE
Sbjct: 3 EETQNKYETAESSEVEVQDRGVFDFLGKKKEEEKPQEEVIVTEFEKITVSEE---KKEE- 58
Query: 692 DHEGEKKHSSLLEKLHRSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVKSTD-------- 537
EGEKKH SLLEKLHRSDSSSSSSS EEEG++G EKKKKKKKEKKE K +
Sbjct: 59 --EGEKKH-SLLEKLHRSDSSSSSSS-EEEGEDG-EKKKKKKKEKKEKKKIEEKIEGYHK 113
Query: 536 ENTSVPVE-VDPAHAEEKKGFLDKIKEKLPGHGKKTTEEATTSPPPPPPSVSETTSTPHE 360
E+TSVPVE V+ +EEKKGFL+KIKEKLPGH K +EEA PPPPP + TS+ HE
Sbjct: 114 EDTSVPVEKVEVVESEEKKGFLEKIKEKLPGH--KKSEEAAAPPPPPPAA----TSSEHE 167
Query: 359 G--AEKKGILEKIKEKIPGYHPKTEEEGTKDKENAAH 255
G EKKGILEKIKEK+PGYH KTEEE K+KE+ H
Sbjct: 168 GEAKEKKGILEKIKEKLPGYHSKTEEE--KEKESGGH 202
>emb|CAC35772.3| dhn1 [Populus euramericana]
Length = 225
Score = 203 bits (516), Expect = 4e-51
Identities = 126/223 (56%), Positives = 147/223 (65%), Gaps = 20/223 (8%)
Frame = -3
Query: 866 ENQNKYESGTTEVETQDRGVFDFLGKKKEEEKPQEEVIVTELEKVSVSEEPEKKKEEVDH 687
E + K + VET+DRG+FDFLG KKEEEKPQEEVI T+ E+ EPE K EE +H
Sbjct: 9 EYETKVGEESGAVETKDRGLFDFLG-KKEEEKPQEEVIATDFEEKLQVSEPETKVEE-EH 66
Query: 686 ---EGEKKHSSLLEKLHRSDSSSSSSSDEEEGKEGEEKKKKKK-----KEKKEV------ 549
E E+K +L EKLHRS SSSSSSS +EE + EEKKKKKK KEK ++
Sbjct: 67 KKKEEEEKKPTLFEKLHRSGSSSSSSSSDEEEGDDEEKKKKKKEKRSLKEKMKISGEKGE 126
Query: 548 KSTDENTSVPVEV----DPAHAEEKKGFLDKIKEKLPGHGKKTTEEATTSPPPPPPSVSE 381
+ E+TSVPVEV P E+KKGFLDKIKEKLPGH K A PPP P VS
Sbjct: 127 EKEHEDTSVPVEVVHTETPHEPEDKKGFLDKIKEKLPGHKK-----ADEVPPPAPEHVSP 181
Query: 380 TTSTPHEG--AEKKGILEKIKEKIPGYHPKTEEEGTKDKENAA 258
+ HEG EKKG+LEKIKEKIPGYHPKTEEE K+KE+A+
Sbjct: 182 EAAVSHEGDAKEKKGLLEKIKEKIPGYHPKTEEEKEKEKESAS 224
>pir||S50766 dehydrin-related protein - garden pea gi|18375|emb|CAA78515.1|
dehydrin-cognate [Pisum sativum]
Length = 216
Score = 201 bits (512), Expect = 1e-50
Identities = 129/218 (59%), Positives = 154/218 (70%), Gaps = 16/218 (7%)
Frame = -3
Query: 869 KENQNKYESGTT----EVETQDRGVFDFLG--KKKEEEKP-QEEVIVTEL-EKVSVSEEP 714
+ENQNKYE T+ E E +DRGVFDFLG KK EE KP Q+E I E KV++ E P
Sbjct: 3 EENQNKYEETTSATNSETEIKDRGVFDFLGGKKKDEEHKPAQDEAIAHEFGHKVTLYEAP 62
Query: 713 EKKKEEVDHEGEKKHSSLLEKLHRSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVKSTDE 534
+ K V+ EGEKKH+SLLEKLHRSDSSSSSSS EEEG++GE++KKKK+K+KK E
Sbjct: 63 SETK--VEEEGEKKHTSLLEKLHRSDSSSSSSS-EEEGEDGEKRKKKKEKKKK------E 113
Query: 533 NTSVPVE-----VDPAHAEEKKGFLDKIKEKLPGHGKKTTEEATTSPP---PPPPSVSET 378
+TSVPVE EE+KGFLDKIKEKLPG G K TEE TT PP P+ +
Sbjct: 114 DTSVPVEKVEVVEGTTGTEEEKGFLDKIKEKLPG-GHKKTEEVTTPPPVVAAHVPTETTA 172
Query: 377 TSTPHEGAEKKGILEKIKEKIPGYHPKTEEEGTKDKEN 264
T+T HEG EKKGILEKIKEK+PGYH KT +G +DK++
Sbjct: 173 TTTIHEG-EKKGILEKIKEKLPGYHAKTATDG-EDKDH 208
>gb|AAN78125.1| dehydrin [Citrus x paradisi]
Length = 234
Score = 195 bits (495), Expect = 1e-48
Identities = 123/234 (52%), Positives = 153/234 (64%), Gaps = 28/234 (11%)
Frame = -3
Query: 872 RKENQNKYESGT-TE--VETQDRGVFDFLGKKKEEEKPQ---EEVIVTELEKVSVSE-EP 714
+++ ++YE TE VET+DRG+ DFLGKK EEEKPQ +EVI TE EKV VSE +P
Sbjct: 7 KQQKSHEYEPSVGTEGAVETKDRGMLDFLGKK-EEEKPQHHDQEVIATEFEKVHVSEPQP 65
Query: 713 EKKKEEVDHEGEKKHSSLLEKLHRSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVKST-- 540
+ ++ + + E+K L+KLHRS SSSSSSSDEEEG + E+KKKKKKEKK +K
Sbjct: 66 KVEEHRKEEKEEEKKPGFLDKLHRSTSSSSSSSDEEEGDD--EEKKKKKKEKKGLKEKLK 123
Query: 539 -------DENTSVPVEV----------DPAHAEEKKGFLDKIKEKLPGHGKKTTEEATTS 411
+E+T+VPVE + AH EEKKGFL+KIKEKLPG KK + S
Sbjct: 124 EKISGEKEEDTTVPVEKLDDVHAPHHQEEAHPEEKKGFLNKIKEKLPGQQKKPGDHQVPS 183
Query: 410 PPPP--PPSVSETTSTPHEGAEKKGILEKIKEKIPGYHPKTEEEGTKDKENAAH 255
PP P SV + E EKKGILEK+KEK+PGYHPK+E+E KDKE AAH
Sbjct: 184 PPAAEHPTSVEAPEA---EAKEKKGILEKLKEKLPGYHPKSEDEKDKDKETAAH 234
>gb|AAF01465.2|AF190474_1 bdn1 [Boea crassifolia]
Length = 252
Score = 189 bits (481), Expect = 4e-47
Identities = 124/235 (52%), Positives = 143/235 (60%), Gaps = 35/235 (14%)
Frame = -3
Query: 854 KYESGTTEVETQDRGVFDFLGKKKEEE---KPQEEVIVTELE-------------KVSVS 723
K E VE DRG+FDF+GKKK+EE K E +E + K V
Sbjct: 21 KIEEIDQPVEASDRGLFDFIGKKKDEEEEKKCDEGKFASEFDDKVQICDEKKEEPKFEVY 80
Query: 722 EEPEKKKEEVDHEGEKKHSSLLEKLHRSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVK- 546
E+P+ + E E EKKH SLLEKLHRS+SSSSSSSDEEE +EG EKKKKKK K +VK
Sbjct: 81 EDPKLEVSEEPKEEEKKHESLLEKLHRSNSSSSSSSDEEEVEEGGEKKKKKKGLKDKVKE 140
Query: 545 ------------STDENTSVPVE-VDPAHA---EEKKGFLDKIKEKLPGHGKKTTEEATT 414
+ E TSVPVE D H EEKKGFLDKIKEKLPG GKKT E
Sbjct: 141 KITGDKKEEAAETKCEETSVPVEKYDEIHTLEPEEKKGFLDKIKEKLPG-GKKT--EEVY 197
Query: 413 SPPPPPPSVSETTSTPH-EGAEKKGILEKIKEKIPGYHPKTEEEGTKDK-ENAAH 255
+PPPP V+E ++ P EG EKKG L+KIKEK+PGYHPK EEE K+K E A H
Sbjct: 198 APPPPKEDVAEYSTAPEAEGKEKKGFLDKIKEKLPGYHPKAEEEKEKEKREEACH 252
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 869,655,822
Number of Sequences: 1393205
Number of extensions: 22333590
Number of successful extensions: 301398
Number of sequences better than 10.0: 7261
Number of HSP's better than 10.0 without gapping: 134601
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 226382
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 55465006400
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)