Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004215A_C01 KMC004215A_c01
(669 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
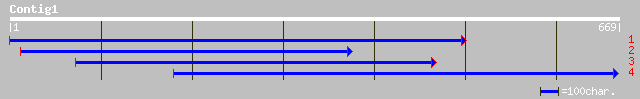
ref|NP_176739.1| unknown protein; protein id: At1g65620.1, suppo... 78 9e-14
dbj|BAB92647.1| B1099D03.24 [Oryza sativa (japonica cultivar-gro... 45 0.001
gb|EAA01825.1| ebiP8498 [Anopheles gambiae str. PEST] 40 0.022
ref|NP_611556.2| CG30389-PC [Drosophila melanogaster] gi|2465674... 39 0.064
ref|NP_725091.1| CG13188-PA [Drosophila melanogaster] gi|2324032... 37 0.18
>ref|NP_176739.1| unknown protein; protein id: At1g65620.1, supported by cDNA:
gi_17227155, supported by cDNA: gi_19918970 [Arabidopsis
thaliana] gi|25404562|pir||C96681 hypothetical protein
F5I14.15 [imported] - Arabidopsis thaliana
gi|2190548|gb|AAB60912.1| EST gb|ATTS1121 comes from
this gene. [Arabidopsis thaliana]
gi|17227156|gb|AAL38032.1|AF447887_1 LOB DOMAIN 6
[Arabidopsis thaliana] gi|19918971|dbj|BAB88693.1|
ASYMMETRIC LEAVES2 [Arabidopsis thaliana]
Length = 199
Score = 78.2 bits (191), Expect = 9e-14
Identities = 58/152 (38%), Positives = 73/152 (47%), Gaps = 3/152 (1%)
Frame = -3
Query: 667 CVGVISILHHQLRKLQMDLLLAKNELSKIQNFGLAMAGQNLAATTAAAAAANMAATAAAT 488
CVGVIS+L HQLR+LQ+DL AK+ELSK Q+ G+ A A AA+ ATA
Sbjct: 84 CVGVISLLQHQLRQLQIDLSCAKSELSKYQSLGILAATHQSLGINLLAGAADGTATAVRD 143
Query: 487 TIPQHMTNMSGGGIGGSGWDNYLHYHHHRHQFFPRDVV---SNFDSGSSYEANPLAINNT 317
HYHH HQFFPR+ + + +G++Y+ LAI
Sbjct: 144 -----------------------HYHH--HQFFPREQMFGGLDVPAGNNYDGGILAIG-- 176
Query: 316 SQQLSSFVQLNQLQLPSAAASVDNRLTTVHPS 221
Q+ Q Q P AAA D R TV PS
Sbjct: 177 --------QITQFQQPRAAAGDDGR-RTVDPS 199
>dbj|BAB92647.1| B1099D03.24 [Oryza sativa (japonica cultivar-group)]
Length = 269
Score = 44.7 bits (104), Expect = 0.001
Identities = 31/87 (35%), Positives = 41/87 (46%)
Frame = -3
Query: 667 CVGVISILHHQLRKLQMDLLLAKNELSKIQNFGLAMAGQNLAATTAAAAAANMAATAAAT 488
CV +ISIL LR+LQ DL AK ELSK Q AAAAAA +A+
Sbjct: 113 CVAIISILQRNLRQLQQDLARAKFELSKYQQ--------------AAAAAAAASASTGTN 158
Query: 487 TIPQHMTNMSGGGIGGSGWDNYLHYHH 407
P M G + +G ++++ H
Sbjct: 159 NGPHSMAEFIGNAV-PNGAQSFINVGH 184
>gb|EAA01825.1| ebiP8498 [Anopheles gambiae str. PEST]
Length = 1186
Score = 40.4 bits (93), Expect = 0.022
Identities = 30/122 (24%), Positives = 54/122 (43%), Gaps = 18/122 (14%)
Frame = -3
Query: 565 AMAGQNLAATTAAAAAANMA------ATAAATTIPQHMTNMSGGGIGGSGWDNYLHYHHH 404
A+AG + +A ++++A A A++++ N S G GG+G + LH+HHH
Sbjct: 220 ALAGNSSSAGCSSSSAGPSAPFGLGCGVASSSSSSISGNNCSPAGSGGNGGNLLLHHHHH 279
Query: 403 RHQFFPRDVVSNFDSGSSYEANPLA------------INNTSQQLSSFVQLNQLQLPSAA 260
Q + + S Y+ N A +N+++ +S Q P+AA
Sbjct: 280 HQQQQQQQQQQQYQSSGGYDPNAAAYMLPARKRPRRTYSNSNEVGTSMAAATMPQQPTAA 339
Query: 259 AS 254
A+
Sbjct: 340 AA 341
>ref|NP_611556.2| CG30389-PC [Drosophila melanogaster] gi|24656749|ref|NP_611555.2|
CG30389-PA [Drosophila melanogaster]
gi|21645182|gb|AAF46685.2| CG30389-PA [Drosophila
melanogaster] gi|21645183|gb|AAM70856.1| CG30389-PC
[Drosophila melanogaster]
Length = 980
Score = 38.9 bits (89), Expect = 0.064
Identities = 26/82 (31%), Positives = 36/82 (43%)
Frame = -3
Query: 547 LAATTAAAAAANMAATAAATTIPQHMTNMSGGGIGGSGWDNYLHYHHHRHQFFPRDVVSN 368
L A++ A AA + ATA + G G S N H+HHH H + SN
Sbjct: 235 LVASSGATAANGVLATATQAQNHHEHHHSGGAGASSSSSSNSNHHHHHHHNNSSGNSGSN 294
Query: 367 FDSGSSYEANPLAINNTSQQLS 302
+S SS + N+TS+ S
Sbjct: 295 HNSNSS--SGGAKDNSTSESAS 314
>ref|NP_725091.1| CG13188-PA [Drosophila melanogaster] gi|23240327|gb|AAF58603.2|
CG13188-PA [Drosophila melanogaster]
Length = 1025
Score = 37.4 bits (85), Expect = 0.18
Identities = 24/61 (39%), Positives = 33/61 (53%)
Frame = -3
Query: 616 DLLLAKNELSKIQNFGLAMAGQNLAATTAAAAAANMAATAAATTIPQHMTNMSGGGIGGS 437
D+L +KN + +N +A A N AA AA+AA AA A A + T+ SGG G+
Sbjct: 880 DMLKSKNYFNS-KNKSIAAAAANAAAAAAASAATTAAAKATAASASIATTSGSGGAEPGA 938
Query: 436 G 434
G
Sbjct: 939 G 939
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 605,590,262
Number of Sequences: 1393205
Number of extensions: 14029390
Number of successful extensions: 81427
Number of sequences better than 10.0: 134
Number of HSP's better than 10.0 without gapping: 58441
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76421
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)