Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004211A_C01 KMC004211A_c01
(602 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
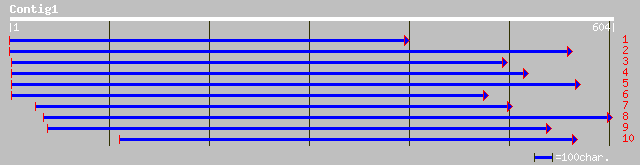
ref|NP_189085.1| unknown protein; protein id: At3g24420.1 [Arabi... 92 5e-18
gb|AAM65892.1| unknown [Arabidopsis thaliana] 63 3e-09
ref|NP_566220.1| expressed protein; protein id: At3g03990.1, sup... 62 4e-09
dbj|BAB91828.1| B1103C09.18 [Oryza sativa (japonica cultivar-gro... 60 2e-08
ref|NP_195463.1| putative protein; protein id: At4g37470.1, supp... 56 4e-07
>ref|NP_189085.1| unknown protein; protein id: At3g24420.1 [Arabidopsis thaliana]
gi|11994705|dbj|BAB02943.1| hydrolase-like protein
[Arabidopsis thaliana]
Length = 273
Score = 92.0 bits (227), Expect = 5e-18
Identities = 44/88 (50%), Positives = 66/88 (75%), Gaps = 1/88 (1%)
Frame = -2
Query: 601 RMRGEVPVSLAKTVFCCDYRDILEKVETACTIIQSSNDMVVPYSVALYMEKKIKGKVTLE 422
+M+ E ++LAK VF D R+IL +V C +IQ ND+VVP SVA +M++KIKGK T+E
Sbjct: 186 KMKPETALALAKIVFGSDEREILGQVSVPCHVIQPGNDVVVPVSVAYFMQEKIKGKSTVE 245
Query: 421 VI-DTVGHFPQLTAHLQLIDLIKGVLGF 341
+I D +GHFPQ+T+HL+L+ +++ +L F
Sbjct: 246 IIEDAIGHFPQMTSHLELLGVMRRLLEF 273
>gb|AAM65892.1| unknown [Arabidopsis thaliana]
Length = 267
Score = 63.2 bits (152), Expect = 3e-09
Identities = 31/84 (36%), Positives = 50/84 (58%)
Frame = -2
Query: 598 MRGEVPVSLAKTVFCCDYRDILEKVETACTIIQSSNDMVVPYSVALYMEKKIKGKVTLEV 419
MR ++ + +++TVF D R +L V +IQ++ D+ VP SVA Y+ + G+ T+E
Sbjct: 182 MRPDISLFVSRTVFNSDLRGVLGLVRVPTCVIQTAKDVSVPASVAEYLRSHLGGETTVET 241
Query: 418 IDTVGHFPQLTAHLQLIDLIKGVL 347
+ T GH PQL+A QL ++ L
Sbjct: 242 LKTEGHLPQLSAPAQLAQFLRRAL 265
>ref|NP_566220.1| expressed protein; protein id: At3g03990.1, supported by cDNA:
6052., supported by cDNA: gi_17381266, supported by
cDNA: gi_20453358 [Arabidopsis thaliana]
gi|6223644|gb|AAF05858.1|AC011698_9 unknown protein
[Arabidopsis thaliana] gi|17381267|gb|AAL36052.1|
AT3g03990/T11I18_10 [Arabidopsis thaliana]
gi|20453359|gb|AAM19918.1| AT3g03990/T11I18_10
[Arabidopsis thaliana]
Length = 267
Score = 62.4 bits (150), Expect = 4e-09
Identities = 31/84 (36%), Positives = 49/84 (57%)
Frame = -2
Query: 598 MRGEVPVSLAKTVFCCDYRDILEKVETACTIIQSSNDMVVPYSVALYMEKKIKGKVTLEV 419
MR ++ + +++TVF D R +L V +IQ++ D+ VP SVA Y+ + G T+E
Sbjct: 182 MRPDISLFVSRTVFNSDLRGVLGLVRVPTCVIQTAKDVSVPASVAEYLRSHLGGDTTVET 241
Query: 418 IDTVGHFPQLTAHLQLIDLIKGVL 347
+ T GH PQL+A QL ++ L
Sbjct: 242 LKTEGHLPQLSAPAQLAQFLRRAL 265
>dbj|BAB91828.1| B1103C09.18 [Oryza sativa (japonica cultivar-group)]
gi|20804634|dbj|BAB92324.1| P0451D05.26 [Oryza sativa
(japonica cultivar-group)]
Length = 302
Score = 60.1 bits (144), Expect = 2e-08
Identities = 31/84 (36%), Positives = 48/84 (56%)
Frame = -2
Query: 598 MRGEVPVSLAKTVFCCDYRDILEKVETACTIIQSSNDMVVPYSVALYMEKKIKGKVTLEV 419
M V +LA+ +F D R +L +V CT++ +S D P V YME +I G+ L
Sbjct: 181 MDPRVADALARMIFLGDNRGVLGRVAAPCTLVHASGDPAAPPCVGRYMEGRI-GRAALVT 239
Query: 418 IDTVGHFPQLTAHLQLIDLIKGVL 347
+D+ GHFPQL A +++ ++ VL
Sbjct: 240 VDSAGHFPQLVAPDEMLRILDAVL 263
>ref|NP_195463.1| putative protein; protein id: At4g37470.1, supported by cDNA:
gi_15810302, supported by cDNA: gi_20259140 [Arabidopsis
thaliana] gi|7450663|pir||T04741 hypothetical protein
F6G17.120 - Arabidopsis thaliana
gi|4468813|emb|CAB38214.1| putative protein [Arabidopsis
thaliana] gi|7270729|emb|CAB80412.1| putative protein
[Arabidopsis thaliana] gi|15810303|gb|AAL07039.1|
unknown protein [Arabidopsis thaliana]
gi|20259141|gb|AAM14286.1| unknown protein [Arabidopsis
thaliana]
Length = 270
Score = 55.8 bits (133), Expect = 4e-07
Identities = 29/80 (36%), Positives = 47/80 (58%)
Frame = -2
Query: 598 MRGEVPVSLAKTVFCCDYRDILEKVETACTIIQSSNDMVVPYSVALYMEKKIKGKVTLEV 419
MR ++ +S+ +T+F D R IL V C I+QS D+ VP V+ Y+ + + +EV
Sbjct: 181 MRPDIALSVGQTIFQSDMRQILPFVTVPCHILQSVKDLAVPVVVSEYLHANLGCESVVEV 240
Query: 418 IDTVGHFPQLTAHLQLIDLI 359
I + GH PQL++ +I +I
Sbjct: 241 IPSDGHLPQLSSPDSVIPVI 260
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,235,612
Number of Sequences: 1393205
Number of extensions: 9980785
Number of successful extensions: 23448
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 22740
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23436
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23711793746
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)