Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004202A_C01 KMC004202A_c01
(614 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
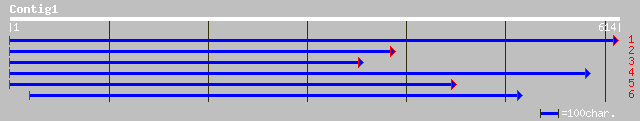
pir||S29851 protein kinase 6 (EC 2.7.1.-) - soybean gi|170047|gb... 172 2e-42
emb|CAA66149.1| PKF1 [Fagus sylvatica] 154 9e-37
emb|CAC09580.1| protein kinase (PK) [Fagus sylvatica] 153 2e-36
ref|NP_568893.1| protein kinase 6 - like; protein id: At5g58950.... 123 2e-27
gb|AAL58946.1|AF462860_1 AT5g58950/k19m22_150 [Arabidopsis thali... 122 4e-27
>pir||S29851 protein kinase 6 (EC 2.7.1.-) - soybean gi|170047|gb|AAA34002.1|
protein kinase gi|444789|prf||1908223A protein kinase
Length = 462
Score = 172 bits (437), Expect = 2e-42
Identities = 78/91 (85%), Positives = 86/91 (93%)
Frame = -2
Query: 487 QNSRPVIPSNCPPAMRALIEQCWSLNPDKRPEFWQVVKVLEQFESSLARDGTLTLVQNPC 308
+NSRP+IPSNCPPAMRALIEQCWSL PDKRPEFWQVVK+LEQFESSLA DGTL+LV NPC
Sbjct: 373 KNSRPIIPSNCPPAMRALIEQCWSLQPDKRPEFWQVVKILEQFESSLASDGTLSLVPNPC 432
Query: 307 SQDHKKGLLHWIQKLGPVHQSNGPMPKPKFT 215
DHKKGLLHWIQKLGP+HQ++GP+PKPKFT
Sbjct: 433 -WDHKKGLLHWIQKLGPLHQNSGPVPKPKFT 462
>emb|CAA66149.1| PKF1 [Fagus sylvatica]
Length = 204
Score = 154 bits (389), Expect = 9e-37
Identities = 74/91 (81%), Positives = 78/91 (85%)
Frame = -2
Query: 487 QNSRPVIPSNCPPAMRALIEQCWSLNPDKRPEFWQVVKVLEQFESSLARDGTLTLVQNPC 308
+N RPVIP +CPPAMRALIEQCWSL +KRPEFWQVVKVLEQFESSLARDGTL LVQ+
Sbjct: 115 KNLRPVIPRDCPPAMRALIEQCWSLQSEKRPEFWQVVKVLEQFESSLARDGTLNLVQSLT 174
Query: 307 SQDHKKGLLHWIQKLGPVHQSNGPMPKPKFT 215
QDHKKGLLHWI KLGPVH NG MPKPK T
Sbjct: 175 CQDHKKGLLHWIHKLGPVH-PNGSMPKPKLT 204
>emb|CAC09580.1| protein kinase (PK) [Fagus sylvatica]
Length = 480
Score = 153 bits (386), Expect = 2e-36
Identities = 74/91 (81%), Positives = 77/91 (84%)
Frame = -2
Query: 487 QNSRPVIPSNCPPAMRALIEQCWSLNPDKRPEFWQVVKVLEQFESSLARDGTLTLVQNPC 308
+N RPVIP CPPAMRALIEQCWSL +KRPEFWQVVKVLEQFESSLARDGTL LVQ+
Sbjct: 391 KNLRPVIPRYCPPAMRALIEQCWSLQSEKRPEFWQVVKVLEQFESSLARDGTLNLVQSLT 450
Query: 307 SQDHKKGLLHWIQKLGPVHQSNGPMPKPKFT 215
QDHKKGLLHWI KLGPVH NG MPKPK T
Sbjct: 451 CQDHKKGLLHWIHKLGPVH-PNGSMPKPKLT 480
>ref|NP_568893.1| protein kinase 6 - like; protein id: At5g58950.1, supported by
cDNA: gi_18087632, supported by cDNA: gi_20260119
[Arabidopsis thaliana] gi|9759226|dbj|BAB09638.1|
protein-tyrosine kinase [Arabidopsis thaliana]
gi|20260120|gb|AAM12958.1| protein-tyrosine kinase
[Arabidopsis thaliana]
Length = 525
Score = 123 bits (309), Expect = 2e-27
Identities = 59/101 (58%), Positives = 69/101 (67%), Gaps = 11/101 (10%)
Frame = -2
Query: 487 QNSRPVIPSNCPPAMRALIEQCWSLNPDKRPEFWQVVKVLEQFESSLARDGTLTLVQNPC 308
+N RP IP +CP AM+ALIEQCWS+ PDKRPEFWQ+VKVLEQF SL R+G L L +
Sbjct: 424 KNIRPAIPGDCPVAMKALIEQCWSVAPDKRPEFWQIVKVLEQFAISLEREGNLNLSSSKI 483
Query: 307 SQDHKKGLLHWIQKLGPVHQSNG-----------PMPKPKF 218
+D +KGL HWIQKLGPVH G +PKPKF
Sbjct: 484 CKDPRKGLKHWIQKLGPVHAGGGGGSSSSGLGGSALPKPKF 524
>gb|AAL58946.1|AF462860_1 AT5g58950/k19m22_150 [Arabidopsis thaliana]
Length = 525
Score = 122 bits (306), Expect = 4e-27
Identities = 59/101 (58%), Positives = 68/101 (66%), Gaps = 11/101 (10%)
Frame = -2
Query: 487 QNSRPVIPSNCPPAMRALIEQCWSLNPDKRPEFWQVVKVLEQFESSLARDGTLTLVQNPC 308
+N RP IP +CP AM+ALIEQCWS+ PDKRPEFWQ+VKVLEQF SL R G L L +
Sbjct: 424 KNIRPAIPGDCPVAMKALIEQCWSVAPDKRPEFWQIVKVLEQFAISLERKGNLNLSSSKI 483
Query: 307 SQDHKKGLLHWIQKLGPVHQSNG-----------PMPKPKF 218
+D +KGL HWIQKLGPVH G +PKPKF
Sbjct: 484 CKDPRKGLKHWIQKLGPVHAGGGGGSSSSGLGGSALPKPKF 524
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 495,397,652
Number of Sequences: 1393205
Number of extensions: 10735692
Number of successful extensions: 25710
Number of sequences better than 10.0: 1287
Number of HSP's better than 10.0 without gapping: 24857
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25689
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24854530794
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)