Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004195A_C01 KMC004195A_c01
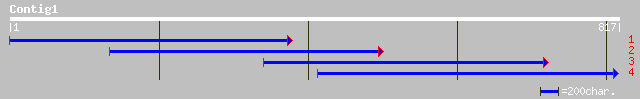
(817 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN41353.1| putative glucose-6-phosphate isomerase [Arabidops... 76 3e-26
gb|AAF24124.1|AF120494_1 phosphoglucose isomerase precursor [Ara... 76 3e-26
ref|NP_194193.1| glucose-6-phosphate isomerase; protein id: At4g... 76 3e-26
pir||T09153 glucose-6-phosphate isomerase (EC 5.3.1.9) precursor... 76 2e-25
ref|ZP_00071859.1| hypothetical protein [Trichodesmium erythraeu... 30 0.45
>gb|AAN41353.1| putative glucose-6-phosphate isomerase [Arabidopsis thaliana]
Length = 613
Score = 75.9 bits (185), Expect(2) = 3e-26
Identities = 33/42 (78%), Positives = 41/42 (97%)
Frame = -3
Query: 815 RVLAVLNEASCQEPVEPLTLEEVAERCHAPPDIEMIYQIIAH 690
RVL+VLNEA+C++PVEPLTLEE+A+RCHAP +IEMIY+IIAH
Sbjct: 538 RVLSVLNEATCKDPVEPLTLEEIADRCHAPEEIEMIYKIIAH 579
Score = 65.5 bits (158), Expect(2) = 3e-26
Identities = 28/39 (71%), Positives = 35/39 (88%)
Frame = -1
Query: 703 KLLLTLAANDRALIAEGSCGSPRSIKVYIGECNVDESYA 587
K++ ++ANDR LIAEG+CGSPRSIKVY+GECNVD+ YA
Sbjct: 575 KIIAHMSANDRVLIAEGNCGSPRSIKVYLGECNVDDLYA 613
>gb|AAF24124.1|AF120494_1 phosphoglucose isomerase precursor [Arabidopsis thaliana]
Length = 612
Score = 75.9 bits (185), Expect(2) = 3e-26
Identities = 33/42 (78%), Positives = 41/42 (97%)
Frame = -3
Query: 815 RVLAVLNEASCQEPVEPLTLEEVAERCHAPPDIEMIYQIIAH 690
RVL+VLNEA+C++PVEPLTLEE+A+RCHAP +IEMIY+IIAH
Sbjct: 537 RVLSVLNEATCKDPVEPLTLEEIADRCHAPEEIEMIYKIIAH 578
Score = 65.5 bits (158), Expect(2) = 3e-26
Identities = 28/39 (71%), Positives = 35/39 (88%)
Frame = -1
Query: 703 KLLLTLAANDRALIAEGSCGSPRSIKVYIGECNVDESYA 587
K++ ++ANDR LIAEG+CGSPRSIKVY+GECNVD+ YA
Sbjct: 574 KIIAHMSANDRVLIAEGNCGSPRSIKVYLGECNVDDLYA 612
>ref|NP_194193.1| glucose-6-phosphate isomerase; protein id: At4g24620.1 [Arabidopsis
thaliana] gi|7484979|pir||T05572 glucose-6-phosphate
isomerase (EC 5.3.1.9) - Arabidopsis thaliana
gi|4220528|emb|CAA23001.1| glucose-6-phosphate isomerase
[Arabidopsis thaliana] gi|7269312|emb|CAB79372.1|
glucose-6-phosphate isomerase [Arabidopsis thaliana]
Length = 611
Score = 75.9 bits (185), Expect(2) = 3e-26
Identities = 33/42 (78%), Positives = 41/42 (97%)
Frame = -3
Query: 815 RVLAVLNEASCQEPVEPLTLEEVAERCHAPPDIEMIYQIIAH 690
RVL+VLNEA+C++PVEPLTLEE+A+RCHAP +IEMIY+IIAH
Sbjct: 536 RVLSVLNEATCKDPVEPLTLEEIADRCHAPEEIEMIYKIIAH 577
Score = 65.5 bits (158), Expect(2) = 3e-26
Identities = 28/39 (71%), Positives = 35/39 (88%)
Frame = -1
Query: 703 KLLLTLAANDRALIAEGSCGSPRSIKVYIGECNVDESYA 587
K++ ++ANDR LIAEG+CGSPRSIKVY+GECNVD+ YA
Sbjct: 573 KIIAHMSANDRVLIAEGNCGSPRSIKVYLGECNVDDLYA 611
>pir||T09153 glucose-6-phosphate isomerase (EC 5.3.1.9) precursor, chloroplast -
spinach gi|3413511|emb|CAA03982.1| glucose-6-phosphate
isomerase [Spinacia oleracea]
Length = 618
Score = 75.9 bits (185), Expect(2) = 2e-25
Identities = 34/42 (80%), Positives = 39/42 (91%)
Frame = -3
Query: 815 RVLAVLNEASCQEPVEPLTLEEVAERCHAPPDIEMIYQIIAH 690
RVLAVLNEASC++PVEPLT+EEVA+ CH P DIEMIY+IIAH
Sbjct: 543 RVLAVLNEASCKDPVEPLTIEEVADHCHCPDDIEMIYKIIAH 584
Score = 62.8 bits (151), Expect(2) = 2e-25
Identities = 26/39 (66%), Positives = 33/39 (83%)
Frame = -1
Query: 703 KLLLTLAANDRALIAEGSCGSPRSIKVYIGECNVDESYA 587
K++ +AANDR ++AEG CGSPRSIK ++GECNVDE YA
Sbjct: 580 KIIAHMAANDRVILAEGDCGSPRSIKAFLGECNVDELYA 618
>ref|ZP_00071859.1| hypothetical protein [Trichodesmium erythraeum IMS101]
Length = 528
Score = 30.4 bits (67), Expect(2) = 0.45
Identities = 13/26 (50%), Positives = 19/26 (73%)
Frame = -3
Query: 767 PLTLEEVAERCHAPPDIEMIYQIIAH 690
PL+LEE++E+ A IE IY+I+ H
Sbjct: 478 PLSLEELSEKAGAGDQIETIYKILRH 503
Score = 25.0 bits (53), Expect(2) = 0.45
Identities = 11/27 (40%), Positives = 16/27 (58%)
Frame = -1
Query: 703 KLLLTLAANDRALIAEGSCGSPRSIKV 623
K+L LAAN R ++ G+ G P + V
Sbjct: 499 KILRHLAANKRGIVFHGNVGQPDKLGV 525
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 642,874,816
Number of Sequences: 1393205
Number of extensions: 13112799
Number of successful extensions: 32262
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 30330
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32149
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42016716300
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)