Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004190A_C01 KMC004190A_c01
(573 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
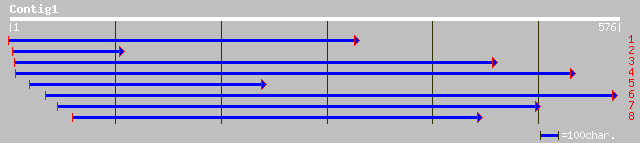
ref|NP_564362.1| unknown protein; protein id: At1g30755.1, suppo... 42 0.007
pir||D86467 protein F23M19.3 [imported] - Arabidopsis thaliana g... 41 0.009
ref|NP_174692.1| hypothetical protein; protein id: At1g34320.1 [... 41 0.009
pir||B86433 hypothetical protein T5I8.20 - Arabidopsis thaliana ... 40 0.021
dbj|BAB89287.1| P0491F11.23 [Oryza sativa (japonica cultivar-gro... 38 0.10
>ref|NP_564362.1| unknown protein; protein id: At1g30755.1, supported by cDNA:
gi_20466801 [Arabidopsis thaliana]
gi|20466802|gb|AAM20718.1| unknown protein [Arabidopsis
thaliana]
Length = 615
Score = 41.6 bits (96), Expect = 0.007
Identities = 33/90 (36%), Positives = 47/90 (51%), Gaps = 2/90 (2%)
Frame = -2
Query: 497 LILSVDSSNKPLGTQLSLEDRRLLEEVIARRRSPGVSKSEELGAANKTKARHLL--RTRS 324
L LS D + K QLSLEDR LL+ V + R P +SKS+EL K K + +RS
Sbjct: 530 LSLSPDFTYK---NQLSLEDRLLLDRVQSIRFGPNLSKSQELVGLKKNKKGFKIWALSRS 586
Query: 323 AGSVPVSEFFGTRFTVSRQNYNTLDIMDGL 234
G+ P + + + + LD++DGL
Sbjct: 587 TGNSPKVDL------SDKNSSSDLDVLDGL 610
>pir||D86467 protein F23M19.3 [imported] - Arabidopsis thaliana
gi|5091616|gb|AAD39604.1|AC007454_3 F23M19.3
[Arabidopsis thaliana]
Length = 609
Score = 41.2 bits (95), Expect = 0.009
Identities = 33/107 (30%), Positives = 55/107 (50%), Gaps = 4/107 (3%)
Frame = -2
Query: 548 RSPPKGHDEFQSKMRQFLILSVDSSNKPLGTQL-SLEDRRLLEEVIARRRSPGVSKSEEL 372
RSP K +S ++ + LS S N +G L + ED+ +L +V RR++PG+SKS+E
Sbjct: 484 RSPVKS--PIRSPNQKTIQLSSGSHNPSMGLPLLTTEDQEMLRDVSKRRKTPGISKSQEF 541
Query: 371 GAANKTK--ARHLLRTRSAGSVPVSEFF-GTRFTVSRQNYNTLDIMD 240
K + H L S+ S + E + T S + +++ I+D
Sbjct: 542 ETVAKARLCKHHRLSKSSSHSPMMGEMMKNKKDTFSTRRPSSVPIID 588
>ref|NP_174692.1| hypothetical protein; protein id: At1g34320.1 [Arabidopsis
thaliana]
Length = 657
Score = 41.2 bits (95), Expect = 0.009
Identities = 33/107 (30%), Positives = 55/107 (50%), Gaps = 4/107 (3%)
Frame = -2
Query: 548 RSPPKGHDEFQSKMRQFLILSVDSSNKPLGTQL-SLEDRRLLEEVIARRRSPGVSKSEEL 372
RSP K +S ++ + LS S N +G L + ED+ +L +V RR++PG+SKS+E
Sbjct: 532 RSPVKS--PIRSPNQKTIQLSSGSHNPSMGLPLLTTEDQEMLRDVSKRRKTPGISKSQEF 589
Query: 371 GAANKTK--ARHLLRTRSAGSVPVSEFF-GTRFTVSRQNYNTLDIMD 240
K + H L S+ S + E + T S + +++ I+D
Sbjct: 590 ETVAKARLCKHHRLSKSSSHSPMMGEMMKNKKDTFSTRRPSSVPIID 636
>pir||B86433 hypothetical protein T5I8.20 - Arabidopsis thaliana
gi|4587531|gb|AAD25762.1|AC007060_20 EST gb|T43746 comes
from this gene. [Arabidopsis thaliana]
Length = 319
Score = 40.0 bits (92), Expect = 0.021
Identities = 32/89 (35%), Positives = 46/89 (50%), Gaps = 2/89 (2%)
Frame = -2
Query: 497 LILSVDSSNKPLGTQLSLEDRRLLEEVIARRRSPGVSKSEELGAANKTKARHLL--RTRS 324
L LS D + K QLSLEDR LL+ V + R P +SKS+EL K K + +RS
Sbjct: 32 LSLSPDFTYK---NQLSLEDRLLLDRVQSIRFGPNLSKSQELVGLKKNKKGFKIWALSRS 88
Query: 323 AGSVPVSEFFGTRFTVSRQNYNTLDIMDG 237
G+ P + + + + LD++DG
Sbjct: 89 TGNSPKVDL------SDKNSSSDLDVLDG 111
>dbj|BAB89287.1| P0491F11.23 [Oryza sativa (japonica cultivar-group)]
Length = 617
Score = 37.7 bits (86), Expect = 0.10
Identities = 30/85 (35%), Positives = 46/85 (53%), Gaps = 1/85 (1%)
Frame = -2
Query: 566 PMPMPSRSPP-KGHDEFQSKMRQFLILSVDSSNKPLGTQLSLEDRRLLEEVIARRRSPGV 390
P+ P RSP +GH S + S++ PL TQ ED+ +L +V R+ PG+
Sbjct: 498 PIKSPVRSPTQRGHTITLSPNK------ASSNSSPLLTQ---EDQDMLRDVKYRKFIPGI 548
Query: 389 SKSEELGAANKTKARHLLRTRSAGS 315
SKS+E +TK+RH ++R + S
Sbjct: 549 SKSQEF----ETKSRHSKQSRLSKS 569
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,882,664
Number of Sequences: 1393205
Number of extensions: 9326092
Number of successful extensions: 23333
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 22650
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23327
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)