Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004184A_C01 KMC004184A_c01
(622 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
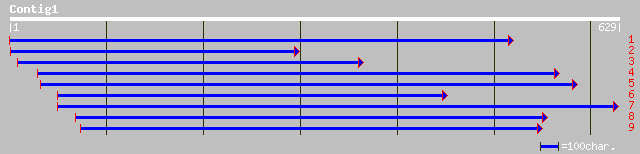
gb|AAM65484.1| unknown [Arabidopsis thaliana] 114 1e-24
ref|NP_567477.1| expressed protein; protein id: At4g15830.1, sup... 114 1e-24
pir||G71423 hypothetical protein - Arabidopsis thaliana gi|22449... 114 1e-24
gb|AAM61606.1| unknown [Arabidopsis thaliana] 89 4e-17
ref|NP_566138.1| expressed protein; protein id: At3g01450.1, sup... 89 5e-17
>gb|AAM65484.1| unknown [Arabidopsis thaliana]
Length = 296
Score = 114 bits (285), Expect = 1e-24
Identities = 57/112 (50%), Positives = 83/112 (73%), Gaps = 3/112 (2%)
Frame = -1
Query: 616 LPLLKKLKGCVSHTNLRIRAKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRLPE 437
LPLL+KL+ V H+N R+RAKAAVS SNCVSKM V EMEEFGM + ++AAD ++D+LPE
Sbjct: 182 LPLLRKLQSYVRHSNPRVRAKAAVSTSNCVSKMEVNEMEEFGMVLLAQMAADQLSDKLPE 241
Query: 436 ARDAARSVATTVYEALTKDAEVEQ---KMEVWQSFCQSKLQPIHALSILKIV 290
AR+AARS+ +++E T + E ++ K E W+ FC+ + ++A +++KIV
Sbjct: 242 AREAARSMVNSLFEKFTWNEEEDEEGSKQEAWKKFCEKNVTGLNAQAMIKIV 293
>ref|NP_567477.1| expressed protein; protein id: At4g15830.1, supported by cDNA:
39757. [Arabidopsis thaliana]
Length = 296
Score = 114 bits (284), Expect = 1e-24
Identities = 57/112 (50%), Positives = 83/112 (73%), Gaps = 3/112 (2%)
Frame = -1
Query: 616 LPLLKKLKGCVSHTNLRIRAKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRLPE 437
LPLL+KL+ V H+N R+RAKAAVS SNCVSKM V EMEEFGM + ++AAD ++D+LPE
Sbjct: 182 LPLLRKLQSYVRHSNPRVRAKAAVSTSNCVSKMEVNEMEEFGMILLAQMAADQLSDKLPE 241
Query: 436 ARDAARSVATTVYEALTKDAEVEQ---KMEVWQSFCQSKLQPIHALSILKIV 290
AR+AARS+ +++E T + E ++ K E W+ FC+ + ++A +++KIV
Sbjct: 242 AREAARSMVNSLFEKFTWNEEEDEEGSKQEAWKKFCEKNVTGLNAQAMIKIV 293
>pir||G71423 hypothetical protein - Arabidopsis thaliana
gi|2244939|emb|CAB10361.1| hypothetical protein
[Arabidopsis thaliana] gi|7268331|emb|CAB78625.1|
hypothetical protein [Arabidopsis thaliana]
Length = 258
Score = 114 bits (284), Expect = 1e-24
Identities = 57/112 (50%), Positives = 83/112 (73%), Gaps = 3/112 (2%)
Frame = -1
Query: 616 LPLLKKLKGCVSHTNLRIRAKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRLPE 437
LPLL+KL+ V H+N R+RAKAAVS SNCVSKM V EMEEFGM + ++AAD ++D+LPE
Sbjct: 144 LPLLRKLQSYVRHSNPRVRAKAAVSTSNCVSKMEVNEMEEFGMILLAQMAADQLSDKLPE 203
Query: 436 ARDAARSVATTVYEALTKDAEVEQ---KMEVWQSFCQSKLQPIHALSILKIV 290
AR+AARS+ +++E T + E ++ K E W+ FC+ + ++A +++KIV
Sbjct: 204 AREAARSMVNSLFEKFTWNEEEDEEGSKQEAWKKFCEKNVTGLNAQAMIKIV 255
>gb|AAM61606.1| unknown [Arabidopsis thaliana]
Length = 326
Score = 89.4 bits (220), Expect = 4e-17
Identities = 45/124 (36%), Positives = 74/124 (59%), Gaps = 5/124 (4%)
Frame = -1
Query: 622 TPLPLLKKLKGCVSHTNLRIRAKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRL 443
+P LL KL+ + + N RIRAKA+ S CV ++G+E + E+G+ ++++ A+ ++D+L
Sbjct: 197 SPALLLPKLQPFLKNRNPRIRAKASTCFSRCVPRLGIEGIREYGIEKLVQAASSQLSDKL 256
Query: 442 PEARDAARSVATTVYEALTKDAEVEQKME-----VWQSFCQSKLQPIHALSILKIVKA*N 278
PE+R+AAR+V + K VE K E WQ FCQS L P+ A +++++
Sbjct: 257 PESREAARAVLLELQTVYKKTTNVEPKEEHPEPVTWQIFCQSNLSPLSAQAVIRVTNVAG 316
Query: 277 WRRE 266
RE
Sbjct: 317 VARE 320
>ref|NP_566138.1| expressed protein; protein id: At3g01450.1, supported by cDNA:
125711. [Arabidopsis thaliana]
gi|6692265|gb|AAF24615.1|AC010870_8 unknown protein
[Arabidopsis thaliana]
Length = 326
Score = 89.0 bits (219), Expect = 5e-17
Identities = 45/124 (36%), Positives = 74/124 (59%), Gaps = 5/124 (4%)
Frame = -1
Query: 622 TPLPLLKKLKGCVSHTNLRIRAKAAVSLSNCVSKMGVEEMEEFGMGEMIEVAADLVNDRL 443
+P LL KL+ + + N RIRAKA+ S CV ++G+E + E+G+ ++++ A+ ++D+L
Sbjct: 197 SPALLLPKLQPFLKNRNPRIRAKASTCFSRCVPRLGIEGIREYGIEKLVQAASSQLSDQL 256
Query: 442 PEARDAARSVATTVYEALTKDAEVEQKME-----VWQSFCQSKLQPIHALSILKIVKA*N 278
PE+R+AAR+V + K VE K E WQ FCQS L P+ A +++++
Sbjct: 257 PESREAARAVLLELQTVYKKTTNVEPKEEHPEPVTWQIFCQSNLSPLSAQAVIRVTNVAG 316
Query: 277 WRRE 266
RE
Sbjct: 317 VARE 320
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,360,332
Number of Sequences: 1393205
Number of extensions: 11562560
Number of successful extensions: 40100
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 35477
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39978
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)