Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004175A_C01 KMC004175A_c01
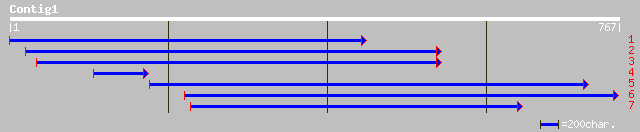
(762 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_188063.1| RNA-binding protein, putative; protein id: At3g... 220 2e-56
ref|NP_175769.1| RNA-binding protein, putative; protein id: At1g... 205 5e-52
ref|NP_174556.1| RNA-binding protein, putative; protein id: At1g... 196 2e-49
gb|AAK53018.1|AF375434_1 At1g32790 [Arabidopsis thaliana] gi|250... 192 6e-48
ref|NP_192799.1| RNA-binding protein RBP37; protein id: At4g1061... 187 1e-46
>ref|NP_188063.1| RNA-binding protein, putative; protein id: At3g14450.1 [Arabidopsis
thaliana] gi|11994215|dbj|BAB01337.1| contains
similarity to RNA-binding protein~gene_id:MOA2.5
[Arabidopsis thaliana]
Length = 327
Score = 220 bits (560), Expect = 2e-56
Identities = 102/124 (82%), Positives = 120/124 (96%)
Frame = -1
Query: 762 TVLGYYPVRVLPSQTAILPVHPTFLPSSDDEREMCSRTVYCTNIDKKISQAEVKNFFESA 583
T+LG+YPVRVLPS+TAILPV+PTFLP S+DEREMC+RT+YCTNIDKK+SQA+V+NFFESA
Sbjct: 202 TMLGFYPVRVLPSKTAILPVNPTFLPRSEDEREMCTRTIYCTNIDKKVSQADVRNFFESA 261
Query: 582 CGEVTRLRLLGDHVHSTRIAFVEFALAESAIVALNCSGMLLGTQPVRVSPSKTPVRPRVT 403
CGEVTRLRLLGD +HSTRIAFVEFALA+SA+ ALNCSGM++G+QP+RVSPSKTPVRPR+T
Sbjct: 262 CGEVTRLRLLGDQLHSTRIAFVEFALADSALSALNCSGMVVGSQPIRVSPSKTPVRPRIT 321
Query: 402 RPTS 391
RP S
Sbjct: 322 RPPS 325
Score = 65.1 bits (157), Expect = 1e-09
Identities = 37/91 (40%), Positives = 51/91 (55%)
Frame = -1
Query: 684 SSDDEREMCSRTVYCTNIDKKISQAEVKNFFESACGEVTRLRLLGDHVHSTRIAFVEFAL 505
S + RTVY ++ID+ +++ + F S CG+V R+ GD R AFVEFA
Sbjct: 131 SKAQREDSIRRTVYVSDIDQSVTEEGLAGLF-SNCGQVVDCRICGDPHSVLRFAFVEFAD 189
Query: 504 AESAIVALNCSGMLLGTQPVRVSPSKTPVRP 412
+ A AL+ G +LG PVRV PSKT + P
Sbjct: 190 DQGAHEALSLGGTMLGFYPVRVLPSKTAILP 220
>ref|NP_175769.1| RNA-binding protein, putative; protein id: At1g53650.1 [Arabidopsis
thaliana] gi|25405657|pir||F96576 probable RNA-binding
protein, 40942-42923 [imported] - Arabidopsis thaliana
gi|12324022|gb|AAG51971.1|AC024260_9 RNA-binding
protein, putative; 40942-42923 [Arabidopsis thaliana]
Length = 314
Score = 205 bits (522), Expect = 5e-52
Identities = 95/124 (76%), Positives = 113/124 (90%)
Frame = -1
Query: 762 TVLGYYPVRVLPSQTAILPVHPTFLPSSDDEREMCSRTVYCTNIDKKISQAEVKNFFESA 583
T++GYYPVRVLPS+TAILPV+PTFLP S+DEREMCSRT+YCTN+DK ++ +V FF+SA
Sbjct: 189 TMIGYYPVRVLPSKTAILPVNPTFLPRSEDEREMCSRTIYCTNVDKNATEDDVNTFFQSA 248
Query: 582 CGEVTRLRLLGDHVHSTRIAFVEFALAESAIVALNCSGMLLGTQPVRVSPSKTPVRPRVT 403
CGEVTRLRLLGD VHSTRIAFVEFA+AESA+ ALNCSG++LG+QP+RVSPSKTPVR R+T
Sbjct: 249 CGEVTRLRLLGDQVHSTRIAFVEFAMAESAVAALNCSGIVLGSQPIRVSPSKTPVRSRIT 308
Query: 402 RPTS 391
R S
Sbjct: 309 RSPS 312
Score = 64.3 bits (155), Expect = 2e-09
Identities = 35/91 (38%), Positives = 52/91 (56%)
Frame = -1
Query: 684 SSDDEREMCSRTVYCTNIDKKISQAEVKNFFESACGEVTRLRLLGDHVHSTRIAFVEFAL 505
S + RTVY ++ID+ +++ + F S+CG+V R+ GD R AFVEF+
Sbjct: 118 SKAQREDSIRRTVYVSDIDQSVTEEGLAGLF-SSCGQVVDCRICGDPNSVLRFAFVEFSD 176
Query: 504 AESAIVALNCSGMLLGTQPVRVSPSKTPVRP 412
+ A AL+ G ++G PVRV PSKT + P
Sbjct: 177 DQGARSALSLGGTMIGYYPVRVLPSKTAILP 207
>ref|NP_174556.1| RNA-binding protein, putative; protein id: At1g32790.1, supported
by cDNA: 40998., supported by cDNA: gi_14030686
[Arabidopsis thaliana] gi|25403416|pir||F86452 protein
F6N18.17 [imported] - Arabidopsis thaliana
gi|6714278|gb|AAF25974.1|AC017118_11 F6N18.17
[Arabidopsis thaliana]
Length = 358
Score = 196 bits (499), Expect = 2e-49
Identities = 92/125 (73%), Positives = 111/125 (88%)
Frame = -1
Query: 762 TVLGYYPVRVLPSQTAILPVHPTFLPSSDDEREMCSRTVYCTNIDKKISQAEVKNFFESA 583
T+LG+YPV+VLPS+TAI PV+PTFLP ++DEREMC+RT+YCTNIDKK++Q++VK FFES
Sbjct: 234 TMLGFYPVKVLPSKTAIAPVNPTFLPRTEDEREMCARTIYCTNIDKKVTQSDVKIFFESF 293
Query: 582 CGEVTRLRLLGDHVHSTRIAFVEFALAESAIVALNCSGMLLGTQPVRVSPSKTPVRPRVT 403
CGEV RLRLLGD+ HSTRIAFVEF +AESAI ALNCSG++LG+ P+RVSPSKTPVRPR
Sbjct: 294 CGEVYRLRLLGDYQHSTRIAFVEFVMAESAIAALNCSGVVLGSLPIRVSPSKTPVRPRSP 353
Query: 402 RPTSH 388
R H
Sbjct: 354 RHPMH 358
Score = 71.2 bits (173), Expect = 1e-11
Identities = 36/91 (39%), Positives = 55/91 (59%)
Frame = -1
Query: 684 SSDDEREMCSRTVYCTNIDKKISQAEVKNFFESACGEVTRLRLLGDHVHSTRIAFVEFAL 505
S ++ RTVY +++D+++++ ++ F S CG+V R+ GD R AF+EF
Sbjct: 163 SMAQREDVIRRTVYVSDLDQQVTEEQLAGLFVS-CGQVVDCRICGDPNSVLRFAFIEFTD 221
Query: 504 AESAIVALNCSGMLLGTQPVRVSPSKTPVRP 412
E A+ ALN SG +LG PV+V PSKT + P
Sbjct: 222 EEGAMTALNLSGTMLGFYPVKVLPSKTAIAP 252
>gb|AAK53018.1|AF375434_1 At1g32790 [Arabidopsis thaliana] gi|25090128|gb|AAN72236.1|
At1g32790/F6N18_9 [Arabidopsis thaliana]
Length = 201
Score = 192 bits (487), Expect = 6e-48
Identities = 90/125 (72%), Positives = 109/125 (87%)
Frame = -1
Query: 762 TVLGYYPVRVLPSQTAILPVHPTFLPSSDDEREMCSRTVYCTNIDKKISQAEVKNFFESA 583
T+LG+YPV+VLPS+TAI PV+PTFLP ++DE EMC+RT+YCTNIDKK++Q++VK FFE
Sbjct: 77 TMLGFYPVKVLPSKTAIAPVNPTFLPRTEDESEMCARTIYCTNIDKKVTQSDVKIFFEYF 136
Query: 582 CGEVTRLRLLGDHVHSTRIAFVEFALAESAIVALNCSGMLLGTQPVRVSPSKTPVRPRVT 403
CGEV RLRLLGD+ HSTRIAFVEF +AESAI ALNCSG++LG+ P+RVSPSKTPVRPR
Sbjct: 137 CGEVYRLRLLGDYQHSTRIAFVEFVMAESAIAALNCSGVVLGSLPIRVSPSKTPVRPRSP 196
Query: 402 RPTSH 388
R H
Sbjct: 197 RHPMH 201
Score = 71.2 bits (173), Expect = 1e-11
Identities = 36/91 (39%), Positives = 55/91 (59%)
Frame = -1
Query: 684 SSDDEREMCSRTVYCTNIDKKISQAEVKNFFESACGEVTRLRLLGDHVHSTRIAFVEFAL 505
S ++ RTVY +++D+++++ ++ F S CG+V R+ GD R AF+EF
Sbjct: 6 SMAQREDVIRRTVYVSDLDQQVTEEQLAGLFVS-CGQVVDCRICGDPNSVLRFAFIEFTD 64
Query: 504 AESAIVALNCSGMLLGTQPVRVSPSKTPVRP 412
E A+ ALN SG +LG PV+V PSKT + P
Sbjct: 65 EEGAMTALNLSGTMLGFYPVKVLPSKTAIAP 95
>ref|NP_192799.1| RNA-binding protein RBP37; protein id: At4g10610.1, supported by
cDNA: 2343., supported by cDNA: gi_1174152, supported by
cDNA: gi_14517527, supported by cDNA: gi_18700203
[Arabidopsis thaliana] gi|7488316|pir||T04196
RNA-binding protein RBP37 - Arabidopsis thaliana
gi|4115925|gb|AAD03436.1| contains similarity to RNA
recognition motifs (Pfam: PF00076, Score=5.5e-23, N=2)
[Arabidopsis thaliana] gi|4539439|emb|CAB40027.1|
RNA-binding protein [Arabidopsis thaliana]
gi|4959384|gb|AAD34325.1| RNA-binding protein
[Arabidopsis thaliana] gi|7267758|emb|CAB78184.1|
RNA-binding protein [Arabidopsis thaliana]
gi|14517528|gb|AAK62654.1| AT4g10610/T4F9_70
[Arabidopsis thaliana] gi|18700204|gb|AAL77712.1|
AT4g10610/T4F9_70 [Arabidopsis thaliana]
gi|21592407|gb|AAM64358.1| RNA-binding protein
[Arabidopsis thaliana]
Length = 336
Score = 187 bits (475), Expect = 1e-46
Identities = 86/118 (72%), Positives = 105/118 (88%)
Frame = -1
Query: 762 TVLGYYPVRVLPSQTAILPVHPTFLPSSDDEREMCSRTVYCTNIDKKISQAEVKNFFESA 583
T+LG+YPV+V+PS+TAI PV+PTFLP ++DEREMC+RT+YCTNIDKK++Q ++K FFES
Sbjct: 211 TMLGFYPVKVMPSKTAIAPVNPTFLPRTEDEREMCARTIYCTNIDKKLTQTDIKLFFESV 270
Query: 582 CGEVTRLRLLGDHVHSTRIAFVEFALAESAIVALNCSGMLLGTQPVRVSPSKTPVRPR 409
CGEV RLRLLGD+ H TRI FVEF +AESAI ALNCSG+LLG+ P+RVSPSKTPVR R
Sbjct: 271 CGEVYRLRLLGDYHHPTRIGFVEFVMAESAIAALNCSGVLLGSLPIRVSPSKTPVRSR 328
Score = 63.2 bits (152), Expect = 4e-09
Identities = 34/85 (40%), Positives = 50/85 (58%)
Frame = -1
Query: 666 EMCSRTVYCTNIDKKISQAEVKNFFESACGEVTRLRLLGDHVHSTRIAFVEFALAESAIV 487
E+ RTVY ++ID+++++ ++ F G+V R+ GD R AF+EF A
Sbjct: 146 EIIRRTVYVSDIDQQVTEEQLAGLF-IGFGQVVDCRICGDPNSVLRFAFIEFTDEVGART 204
Query: 486 ALNCSGMLLGTQPVRVSPSKTPVRP 412
ALN SG +LG PV+V PSKT + P
Sbjct: 205 ALNLSGTMLGFYPVKVMPSKTAIAP 229
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 639,767,212
Number of Sequences: 1393205
Number of extensions: 13366431
Number of successful extensions: 32065
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 31095
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32045
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 36974710344
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)