Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004172A_C01 KMC004172A_c01
(721 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
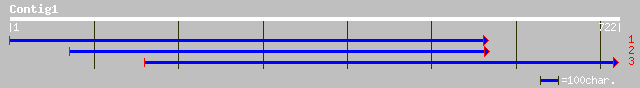
ref|NP_193602.1| extensin-like protein; protein id: At4g18670.1 ... 185 5e-46
ref|NP_189091.1| disease resistance protein, putative; protein i... 180 2e-44
ref|NP_193070.1| extensin-like protein; protein id: At4g13340.1 ... 159 3e-38
ref|NP_188532.1| hypothetical protein; protein id: At3g19020.1 [... 129 4e-29
dbj|BAB01698.1| extensin protein-like [Arabidopsis thaliana] 129 4e-29
>ref|NP_193602.1| extensin-like protein; protein id: At4g18670.1 [Arabidopsis
thaliana] gi|7484957|pir||T04859 extensin homolog
F28A21.80 - Arabidopsis thaliana
gi|4539386|emb|CAB37452.1| extensin-like protein
[Arabidopsis thaliana] gi|7268661|emb|CAB78869.1|
extensin-like protein [Arabidopsis thaliana]
Length = 839
Score = 185 bits (470), Expect = 5e-46
Identities = 87/130 (66%), Positives = 105/130 (79%)
Frame = +3
Query: 330 LLIAVTVTFSATSSAAHRVSDGGLTDAEAMYIKQRQLLYYRDEFGDRGEKVTVDPSLVFE 509
+L+ +T F++ S + +G L+D E I QRQLLY+RDEFGDRGE V VDPSLVFE
Sbjct: 15 VLLFITFFFTSISYSLSLTFNGDLSDNEVRLITQRQLLYFRDEFGDRGENVDVDPSLVFE 74
Query: 510 NNRIRNAYIALQAWKEAILSDPRNYTVNWVGADVCSYSGVFCAPAPDNPKIRTVAGIDLK 689
N R+RNAYIALQAWK+AILSDP N+T NW+G+DVCSY+GV+CAPA DN +IRTVAGIDL
Sbjct: 75 NPRLRNAYIALQAWKQAILSDPNNFTTNWIGSDVCSYTGVYCAPALDNRRIRTVAGIDLN 134
Query: 690 HGDIAGYLPE 719
H DIAGYLP+
Sbjct: 135 HADIAGYLPQ 144
>ref|NP_189091.1| disease resistance protein, putative; protein id: At3g24480.1
[Arabidopsis thaliana] gi|9294099|dbj|BAB01951.1|
extensin-like protein [Arabidopsis thaliana]
Length = 494
Score = 180 bits (456), Expect = 2e-44
Identities = 87/137 (63%), Positives = 108/137 (78%)
Frame = +3
Query: 309 LLILYPILLIAVTVTFSATSSAAHRVSDGGLTDAEAMYIKQRQLLYYRDEFGDRGEKVTV 488
LL+L+ ++ + S +S+A L+D E +I++RQLLYYRDEFGDRGE VTV
Sbjct: 11 LLLLFFFFFFEISHSLSISSNAP-------LSDTEVRFIQRRQLLYYRDEFGDRGENVTV 63
Query: 489 DPSLVFENNRIRNAYIALQAWKEAILSDPRNYTVNWVGADVCSYSGVFCAPAPDNPKIRT 668
DPSL+FEN R+R+AYIALQAWK+AILSDP N TVNW+G++VC+Y+GVFC+ A DN KIRT
Sbjct: 64 DPSLIFENPRLRSAYIALQAWKQAILSDPNNITVNWIGSNVCNYTGVFCSKALDNRKIRT 123
Query: 669 VAGIDLKHGDIAGYLPE 719
VAGIDL H DIAGYLPE
Sbjct: 124 VAGIDLNHADIAGYLPE 140
>ref|NP_193070.1| extensin-like protein; protein id: At4g13340.1 [Arabidopsis
thaliana] gi|7484961|pir||T06291 extensin homolog
T9E8.80 - Arabidopsis thaliana
gi|4584539|emb|CAB40769.1| extensin-like protein
[Arabidopsis thaliana] gi|7268037|emb|CAB78376.1|
extensin-like protein [Arabidopsis thaliana]
Length = 760
Score = 159 bits (403), Expect = 3e-38
Identities = 83/137 (60%), Positives = 103/137 (74%), Gaps = 1/137 (0%)
Frame = +3
Query: 312 LILYPILLIAVTVTFSATSSAAHRVSDGG-LTDAEAMYIKQRQLLYYRDEFGDRGEKVTV 488
++L+ LI +T S +S DGG L+D E +I++RQLL EF +R K+TV
Sbjct: 7 ILLFFFFLINLTNALSISS-------DGGVLSDNEVRHIQRRQLL----EFAERSVKITV 55
Query: 489 DPSLVFENNRIRNAYIALQAWKEAILSDPRNYTVNWVGADVCSYSGVFCAPAPDNPKIRT 668
DPSL FEN R+RNAYIALQAWK+AILSDP N+T NW+G++VC+Y+GVFC+PA DN KIRT
Sbjct: 56 DPSLNFENPRLRNAYIALQAWKQAILSDPNNFTSNWIGSNVCNYTGVFCSPALDNRKIRT 115
Query: 669 VAGIDLKHGDIAGYLPE 719
VAGIDL H DIAGYLPE
Sbjct: 116 VAGIDLNHADIAGYLPE 132
>ref|NP_188532.1| hypothetical protein; protein id: At3g19020.1 [Arabidopsis
thaliana]
Length = 951
Score = 129 bits (324), Expect = 4e-29
Identities = 71/132 (53%), Positives = 85/132 (63%), Gaps = 2/132 (1%)
Frame = +3
Query: 327 ILLIAVTVTFSATSSAAHRVSDGGLTDAEAMYIKQRQLLYYRDEFGDRGEKVT--VDPSL 500
+LL T++ S+AA LTD EA ++ +RQLL E GD + + VD L
Sbjct: 9 LLLFCFTISIFFYSAAA-------LTDEEASFLTRRQLLAL-SENGDLPDDIEYEVDLDL 60
Query: 501 VFENNRIRNAYIALQAWKEAILSDPRNYTVNWVGADVCSYSGVFCAPAPDNPKIRTVAGI 680
F NNR++ AYIALQAWK+A SDP N NWVG DVCSY GVFCAPA D+P + VAGI
Sbjct: 61 KFANNRLKRAYIALQAWKKAFYSDPFNTAANWVGPDVCSYKGVFCAPALDDPSVLVVAGI 120
Query: 681 DLKHGDIAGYLP 716
DL H DIAGYLP
Sbjct: 121 DLNHADIAGYLP 132
>dbj|BAB01698.1| extensin protein-like [Arabidopsis thaliana]
Length = 956
Score = 129 bits (324), Expect = 4e-29
Identities = 71/132 (53%), Positives = 85/132 (63%), Gaps = 2/132 (1%)
Frame = +3
Query: 327 ILLIAVTVTFSATSSAAHRVSDGGLTDAEAMYIKQRQLLYYRDEFGDRGEKVT--VDPSL 500
+LL T++ S+AA LTD EA ++ +RQLL E GD + + VD L
Sbjct: 14 LLLFCFTISIFFYSAAA-------LTDEEASFLTRRQLLAL-SENGDLPDDIEYEVDLDL 65
Query: 501 VFENNRIRNAYIALQAWKEAILSDPRNYTVNWVGADVCSYSGVFCAPAPDNPKIRTVAGI 680
F NNR++ AYIALQAWK+A SDP N NWVG DVCSY GVFCAPA D+P + VAGI
Sbjct: 66 KFANNRLKRAYIALQAWKKAFYSDPFNTAANWVGPDVCSYKGVFCAPALDDPSVLVVAGI 125
Query: 681 DLKHGDIAGYLP 716
DL H DIAGYLP
Sbjct: 126 DLNHADIAGYLP 137
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 657,138,962
Number of Sequences: 1393205
Number of extensions: 15022089
Number of successful extensions: 60874
Number of sequences better than 10.0: 50
Number of HSP's better than 10.0 without gapping: 51166
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 60080
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33499052993
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)