Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004155A_C01 KMC004155A_c01
(615 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
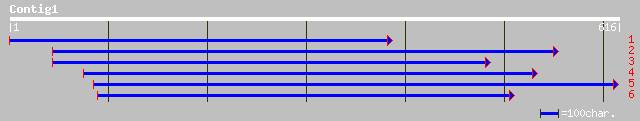
gb|AAC69137.2| putative tyrosyl-tRNA synthetase [Arabidopsis tha... 144 7e-34
ref|NP_565773.2| putative tyrosyl-tRNA synthetase; protein id: A... 144 7e-34
pir||T03741 probable tyrosine-tRNA ligase (EC 6.1.1.1) - common ... 142 3e-33
ref|NP_174157.1| tyrosyl-tRNA synthetase, putative; protein id: ... 129 2e-29
pir||A86410 protein F3M18.22 [imported] - Arabidopsis thaliana g... 129 2e-29
>gb|AAC69137.2| putative tyrosyl-tRNA synthetase [Arabidopsis thaliana]
Length = 351
Score = 144 bits (364), Expect = 7e-34
Identities = 64/92 (69%), Positives = 78/92 (84%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEYIKY+I PWF+EFTVERN + GGNKT+KS E++ A YESGELHPGDLK L +
Sbjct: 260 GNPCLEYIKYIILPWFDEFTVERNEEYGGNKTYKSFEDIAADYESGELHPGDLKKGLMNA 319
Query: 434 LNKILEPVREHFRKDNNAKELLKRVKAYRITK 339
LNKIL+PVR+HF+ D AK LLK++KAYR+T+
Sbjct: 320 LNKILQPVRDHFKTDARAKNLLKQIKAYRVTR 351
>ref|NP_565773.2| putative tyrosyl-tRNA synthetase; protein id: At2g33840.1,
supported by cDNA: gi_18700118 [Arabidopsis thaliana]
gi|18700119|gb|AAL77671.1| At2g33840/T1B8.14
[Arabidopsis thaliana] gi|21464573|gb|AAM52241.1|
At2g33840/T1B8.14 [Arabidopsis thaliana]
Length = 385
Score = 144 bits (364), Expect = 7e-34
Identities = 64/92 (69%), Positives = 78/92 (84%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEYIKY+I PWF+EFTVERN + GGNKT+KS E++ A YESGELHPGDLK L +
Sbjct: 294 GNPCLEYIKYIILPWFDEFTVERNEEYGGNKTYKSFEDIAADYESGELHPGDLKKGLMNA 353
Query: 434 LNKILEPVREHFRKDNNAKELLKRVKAYRITK 339
LNKIL+PVR+HF+ D AK LLK++KAYR+T+
Sbjct: 354 LNKILQPVRDHFKTDARAKNLLKQIKAYRVTR 385
>pir||T03741 probable tyrosine-tRNA ligase (EC 6.1.1.1) - common tobacco
gi|1841468|emb|CAA71881.1| Tyrosyl-tRNA synthetase
[Nicotiana tabacum]
Length = 408
Score = 142 bits (358), Expect = 3e-33
Identities = 59/91 (64%), Positives = 80/91 (87%)
Frame = -2
Query: 611 NPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKSL 432
NPCLEYIKY++ PWFNEF +ER+A+NG +KT+K+ EEL A YESG LHP DLKPAL+K++
Sbjct: 318 NPCLEYIKYIVLPWFNEFKIERSAENGADKTYKNFEELAADYESGNLHPADLKPALSKAI 377
Query: 431 NKILEPVREHFRKDNNAKELLKRVKAYRITK 339
NKIL+PVR+HF+ D AK+L+KRVK++++T+
Sbjct: 378 NKILQPVRDHFKNDQKAKDLMKRVKSFKVTR 408
>ref|NP_174157.1| tyrosyl-tRNA synthetase, putative; protein id: At1g28350.1
[Arabidopsis thaliana]
Length = 866
Score = 129 bits (325), Expect = 2e-29
Identities = 57/91 (62%), Positives = 76/91 (82%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEY+K++I PWF+EFTVER+ GGN+TFKS E++ YESG+LHP DLK AL+K+
Sbjct: 308 GNPCLEYVKHIILPWFSEFTVERDEKYGGNRTFKSFEDITTDYESGQLHPKDLKDALSKA 367
Query: 434 LNKILEPVREHFRKDNNAKELLKRVKAYRIT 342
LNKIL+PVR+HF+ ++ AK LLK+VK +I+
Sbjct: 368 LNKILQPVRDHFKTNSRAKNLLKQVKVIKIS 398
Score = 102 bits (254), Expect = 4e-21
Identities = 57/134 (42%), Positives = 74/134 (54%), Gaps = 42/134 (31%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEY+KY++ P FNEF VE + NGGNKTF S E++VA YE+GELHP DLK AL K+
Sbjct: 734 GNPCLEYVKYIVLPRFNEFKVE-SEKNGGNKTFNSFEDIVADYETGELHPEDLKKALMKA 792
Query: 434 LNKIL------------------------------------------EPVREHFRKDNNA 381
LN L +PVR+HF+ + A
Sbjct: 793 LNITLQFLASVQSSWNMKLVSFQSLLDVRVINFMYMNDMLLIVTVLSQPVRDHFKTNERA 852
Query: 380 KELLKRVKAYRITK 339
K LL++VKA+R+T+
Sbjct: 853 KNLLEQVKAFRVTR 866
>pir||A86410 protein F3M18.22 [imported] - Arabidopsis thaliana
gi|6560759|gb|AAF16759.1|AC010155_12 F3M18.22
[Arabidopsis thaliana]
Length = 895
Score = 129 bits (325), Expect = 2e-29
Identities = 57/91 (62%), Positives = 76/91 (82%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEY+K++I PWF+EFTVER+ GGN+TFKS E++ YESG+LHP DLK AL+K+
Sbjct: 308 GNPCLEYVKHIILPWFSEFTVERDEKYGGNRTFKSFEDITTDYESGQLHPKDLKDALSKA 367
Query: 434 LNKILEPVREHFRKDNNAKELLKRVKAYRIT 342
LNKIL+PVR+HF+ ++ AK LLK+VK +I+
Sbjct: 368 LNKILQPVRDHFKTNSRAKNLLKQVKVIKIS 398
Score = 90.5 bits (223), Expect = 2e-17
Identities = 43/66 (65%), Positives = 52/66 (78%)
Frame = -2
Query: 614 GNPCLEYIKYLIFPWFNEFTVERNADNGGNKTFKSSEELVAAYESGELHPGDLKPALAKS 435
GNPCLEY+KY++ P FNEF VE + NGGNKTF S E++VA YE+GELHP DLK AL K+
Sbjct: 734 GNPCLEYVKYIVLPRFNEFKVE-SEKNGGNKTFNSFEDIVADYETGELHPEDLKKALMKA 792
Query: 434 LNKILE 417
LN L+
Sbjct: 793 LNITLQ 798
Score = 37.7 bits (86), Expect = 0.12
Identities = 14/27 (51%), Positives = 23/27 (84%)
Frame = -2
Query: 419 EPVREHFRKDNNAKELLKRVKAYRITK 339
+PVR+HF+ + AK LL++VKA+R+T+
Sbjct: 869 QPVRDHFKTNERAKNLLEQVKAFRVTR 895
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,695,726
Number of Sequences: 1393205
Number of extensions: 10091412
Number of successful extensions: 26292
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 25582
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26275
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24854530794
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)