Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
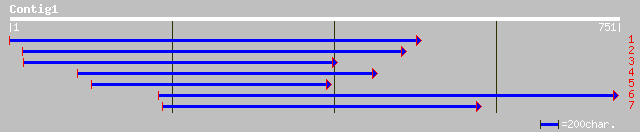
Query= KMC004153A_C01 KMC004153A_c01
(734 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_195694.1| putative protein; protein id: At4g39840.1, supp... 266 2e-70
ref|XP_134237.1| similar to open reading frame (251 AA) [Mus mus... 38 0.13
gb|AAB59264.1| smooth muscle caldesmon 36 0.64
gb|AAA67843.1| l-caldesmon 36 0.64
ref|NP_648931.1| CG11915-PA [Drosophila melanogaster] gi|1676810... 36 0.64
>ref|NP_195694.1| putative protein; protein id: At4g39840.1, supported by cDNA:
gi_18175808 [Arabidopsis thaliana]
gi|7487699|pir||T06090 hypothetical protein T5J17.10 -
Arabidopsis thaliana gi|4490735|emb|CAB38897.1| putative
protein [Arabidopsis thaliana]
gi|7271039|emb|CAB80647.1| putative protein [Arabidopsis
thaliana] gi|18175809|gb|AAL59931.1| unknown protein
[Arabidopsis thaliana] gi|22136848|gb|AAM91768.1|
unknown protein [Arabidopsis thaliana]
Length = 451
Score = 266 bits (680), Expect = 2e-70
Identities = 124/161 (77%), Positives = 144/161 (89%)
Frame = -2
Query: 730 SLVLNKIKAYFSLQKLLLFIQAYLSIYFSILCLSSFVTGLEPLRFFFSTSQSSYLFLQVI 551
SL+ N+ KAYFSLQK+L+FIQ YLSIYFSILCLSS VTG+EPL+F ++TS S+Y+ LQ++
Sbjct: 291 SLIFNRFKAYFSLQKILIFIQIYLSIYFSILCLSSLVTGIEPLKFLYATSSSTYVCLQIL 350
Query: 550 QTLAYVLYLLLLLMYLVLVFSTDSGLGSKFLGLTQTFVGFAVGLHYYMTVFHRVVLRQPP 371
QTL YV YLLLLLMYLVLVFSTD GLG K LGL QTFVGFAVGLHYY+ VFHRVVLRQPP
Sbjct: 351 QTLGYVFYLLLLLMYLVLVFSTDCGLGLKVLGLAQTFVGFAVGLHYYVAVFHRVVLRQPP 410
Query: 370 KTNWKVHGIYATCFFVICVLAGADRRKKSYLEEGGEEGKKN 248
KTNWK+HG+YATCF +IC+L+ A+RRKK YLEEGG+EGKKN
Sbjct: 411 KTNWKIHGVYATCFLLICLLSNAERRKKEYLEEGGDEGKKN 451
>ref|XP_134237.1| similar to open reading frame (251 AA) [Mus musculus]
Length = 279
Score = 38.1 bits (87), Expect = 0.13
Identities = 25/100 (25%), Positives = 46/100 (46%)
Frame = -2
Query: 685 LLLFIQAYLSIYFSILCLSSFVTGLEPLRFFFSTSQSSYLFLQVIQTLAYVLYLLLLLMY 506
L +++ YLSIY S +CLSS + L + S S YL + + Y + L + L
Sbjct: 107 LSIYLSIYLSIYLSYVCLSSIYLSIYHLSIYLSIYLSIYLSIYHVCLSIYHVCLSIYLSI 166
Query: 505 LVLVFSTDSGLGSKFLGLTQTFVGFAVGLHYYMTVFHRVV 386
+ L S +L + V ++ L Y++++ ++
Sbjct: 167 ICL---------SAYLSIYHMSVCLSIYLSMYLSIYLSII 197
Score = 37.0 bits (84), Expect = 0.29
Identities = 28/112 (25%), Positives = 51/112 (45%), Gaps = 1/112 (0%)
Frame = -2
Query: 727 LVLNKIKAYFSLQ-KLLLFIQAYLSIYFSILCLSSFVTGLEPLRFFFSTSQSSYLFLQVI 551
+ L I Y S+ L L + YLSIY S +CLS L + S + L +
Sbjct: 40 ICLLNISIYLSIYLSLSLSLSLYLSIYLSYVCLSIIYLYLSIYLSIYHMSVCLSICLSYV 99
Query: 550 QTLAYVLYLLLLLMYLVLVFSTDSGLGSKFLGLTQTFVGFAVGLHYYMTVFH 395
L+ +YL + L + ++ + L S +L + + ++ L Y++++H
Sbjct: 100 -CLSISIYLSIYLSIYLSIYLSYVCLSSIYLSIYHLSIYLSIYLSIYLSIYH 150
Score = 33.5 bits (75), Expect = 3.2
Identities = 24/74 (32%), Positives = 37/74 (49%), Gaps = 4/74 (5%)
Frame = -2
Query: 712 IKAYFSLQK----LLLFIQAYLSIYFSILCLSSFVTGLEPLRFFFSTSQSSYLFLQVIQT 545
+ AY S+ L +++ YLSIY SI+CL SF+ L + S ++L +I
Sbjct: 169 LSAYLSIYHMSVCLSIYLSMYLSIYLSIICL-SFIYLSVYLSIYLPIICLSIIYLPIICL 227
Query: 544 LAYVLYLLLLLMYL 503
L ++ L MYL
Sbjct: 228 SIIYLPIIYLSMYL 241
Score = 32.0 bits (71), Expect = 9.3
Identities = 24/99 (24%), Positives = 51/99 (51%)
Frame = -2
Query: 697 SLQKLLLFIQAYLSIYFSILCLSSFVTGLEPLRFFFSTSQSSYLFLQVIQTLAYVLYLLL 518
S+ ++ + AYLSIY +CLS +++ L + S S+++L V ++ Y+ + L
Sbjct: 161 SIYLSIICLSAYLSIYHMSVCLSIYLSMY--LSIYLSIICLSFIYLSVYLSI-YLPIICL 217
Query: 517 LLMYLVLVFSTDSGLGSKFLGLTQTFVGFAVGLHYYMTV 401
++YL ++ + L +L + + + V ++ M V
Sbjct: 218 SIIYLPIICLSIIYLPIIYLSMYLSSISLCVCMYICMYV 256
>gb|AAB59264.1| smooth muscle caldesmon
Length = 438
Score = 35.8 bits (81), Expect = 0.64
Identities = 25/105 (23%), Positives = 49/105 (45%)
Frame = +2
Query: 389 HPVKHRHVIM*PDSKPHERLG*AQKLRA*PRVRGENQNKVHEKQEKVENIS*RLDNLQEE 568
H +KH P G AQ+ A R+ + + + E+ E + + E
Sbjct: 150 HKLKHAENTFSPGRSGARPSGDAQEAEAGKRLEELRRRRGETESEEFEKLKQKQQEAALE 209
Query: 569 ITRLRSREEKPQRLKPGNKRREAED*EVDREVSLDEEQELLK*EI 703
+ L+ + E+ +++ ++R+ ++ E DR+V +EE+ LK EI
Sbjct: 210 LEELKKKREERRKVLEEEEQRKKQE-EADRKVREEEEKRRLKEEI 253
>gb|AAA67843.1| l-caldesmon
Length = 533
Score = 35.8 bits (81), Expect = 0.64
Identities = 25/105 (23%), Positives = 49/105 (45%)
Frame = +2
Query: 389 HPVKHRHVIM*PDSKPHERLG*AQKLRA*PRVRGENQNKVHEKQEKVENIS*RLDNLQEE 568
H +KH P G AQ+ A R+ + + + E+ E + + E
Sbjct: 246 HKLKHAENTFSPGRSGARPSGDAQEAEAGKRLEELRRRRGETESEEFEKLKQKQQEAALE 305
Query: 569 ITRLRSREEKPQRLKPGNKRREAED*EVDREVSLDEEQELLK*EI 703
+ L+ + E+ +++ ++R+ ++ E DR+V +EE+ LK EI
Sbjct: 306 LEELKKKREERRKVLEEEEQRKKQE-EADRKVREEEEKRRLKEEI 349
>ref|NP_648931.1| CG11915-PA [Drosophila melanogaster] gi|16768106|gb|AAL28272.1|
GH17145p [Drosophila melanogaster]
gi|23093268|gb|AAF49397.2| CG11915-PA [Drosophila
melanogaster]
Length = 694
Score = 35.8 bits (81), Expect = 0.64
Identities = 24/64 (37%), Positives = 34/64 (52%)
Frame = +2
Query: 491 ENQNKVHEKQEKVENIS*RLDNLQEEITRLRSREEKPQRLKPGNKRREAED*EVDREVSL 670
ENQ K E +EK RL+N + I R REE +RL+ ++RE E E +R + +
Sbjct: 182 ENQRKQRENEEKE-----RLENERRLIDAEREREENERRLQEAEEQRERE--ESERRIVV 234
Query: 671 DEEQ 682
E Q
Sbjct: 235 AERQ 238
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 620,278,831
Number of Sequences: 1393205
Number of extensions: 13896682
Number of successful extensions: 47705
Number of sequences better than 10.0: 82
Number of HSP's better than 10.0 without gapping: 43848
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 47165
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 34906576228
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)