Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004135A_C01 KMC004135A_c01
(530 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
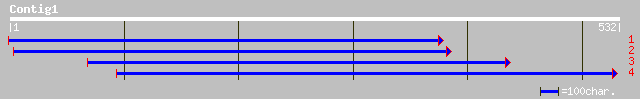
gb|AAL66194.1|AF386512_1 cytochrome P450 [Pyrus communis] 135 3e-31
emb|CAC26932.1| ferulate-5-hydroxylase [Arabidopsis thaliana] gi... 105 3e-22
emb|CAC26936.1| ferulate-5-hydroxylase [Arabidopsis thaliana] gi... 105 3e-22
ref|NP_195345.1| ferulate-5-hydroxylase (FAH1); protein id: At4g... 105 3e-22
gb|AAG14961.1|AF214007_1 cytochrome p450-dependent monooxygenase... 104 6e-22
>gb|AAL66194.1|AF386512_1 cytochrome P450 [Pyrus communis]
Length = 506
Score = 135 bits (340), Expect = 3e-31
Identities = 61/97 (62%), Positives = 71/97 (72%)
Frame = -3
Query: 522 DNAEEFYPERFTNNNFDYRGHDFQFIPFGFGRRGCPGMNLGLASVKLVVAQLVHCFCWEL 343
DNAEEFYPERF N N D RGHDFQ IPFG GRRGCP M LGL +V+L + L+HC WEL
Sbjct: 410 DNAEEFYPERFMNRNVDLRGHDFQLIPFGSGRRGCPAMQLGLTTVRLALGNLLHCSNWEL 469
Query: 342 PSNTTPNELDMSEKSGFSIPRAKHLFAVPTYRLLKKA 232
PS P +LDM+EK G S+ +AKHL A PT RL ++
Sbjct: 470 PSGMLPKDLDMTEKFGLSLSKAKHLLATPTCRLYNES 506
>emb|CAC26932.1| ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578895|emb|CAC26933.1| ferulate-5-hydroxylase
[Arabidopsis thaliana]
Length = 520
Score = 105 bits (262), Expect = 3e-22
Identities = 49/97 (50%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Frame = -3
Query: 528 WSDNAEEFYPERFTNNNF-DYRGHDFQFIPFGFGRRGCPGMNLGLASVKLVVAQLVHCFC 352
W+D + F P RF D++G +F+FIPFG GRR CPGM LGL ++ L VA ++HCF
Sbjct: 422 WTD-PDTFRPSRFLEPGVPDFKGSNFEFIPFGSGRRSCPGMQLGLYALDLAVAHILHCFT 480
Query: 351 WELPSNTTPNELDMSEKSGFSIPRAKHLFAVPTYRLL 241
W+LP P+ELDM++ G + P+A LFAVPT RL+
Sbjct: 481 WKLPDGMKPSELDMNDVFGLTAPKATRLFAVPTTRLI 517
>emb|CAC26936.1| ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578903|emb|CAC26937.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578905|emb|CAC26938.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578907|emb|CAC26939.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578909|emb|CAC26940.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578911|emb|CAC26941.1| ferulate-5-hydroxylase
[Arabidopsis thaliana]
Length = 520
Score = 105 bits (262), Expect = 3e-22
Identities = 49/97 (50%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Frame = -3
Query: 528 WSDNAEEFYPERFTNNNF-DYRGHDFQFIPFGFGRRGCPGMNLGLASVKLVVAQLVHCFC 352
W+D + F P RF D++G +F+FIPFG GRR CPGM LGL ++ L VA ++HCF
Sbjct: 422 WTD-PDTFRPSRFLEPGVPDFKGSNFEFIPFGSGRRSCPGMQLGLYALDLAVAHILHCFT 480
Query: 351 WELPSNTTPNELDMSEKSGFSIPRAKHLFAVPTYRLL 241
W+LP P+ELDM++ G + P+A LFAVPT RL+
Sbjct: 481 WKLPDGMKPSELDMNDVFGLTAPKATRLFAVPTTRLI 517
>ref|NP_195345.1| ferulate-5-hydroxylase (FAH1); protein id: At4g36220.1, supported
by cDNA: gi_1488254 [Arabidopsis thaliana]
gi|5921932|sp|Q42600|C84A_ARATH Cytochrome P450 84A1
(Ferulate-5-hydroxylase) (F5H) gi|7430674|pir||T04591
ferulate-5-hydroxylase (EC 1.-.-.-) - Arabidopsis
thaliana gi|1488255|gb|AAC49389.1|
ferulate-5-hydroxylase gi|2961381|emb|CAA18128.1|
ferulate-5-hydroxylase (FAH1) [Arabidopsis thaliana]
gi|3925365|gb|AAD11580.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|7270575|emb|CAB80293.1|
ferulate-5-hydroxylase (FAH1) [Arabidopsis thaliana]
gi|12578873|emb|CAC26922.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578875|emb|CAC26923.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578877|emb|CAC26924.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578879|emb|CAC26925.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578881|emb|CAC26926.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578883|emb|CAC26927.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578885|emb|CAC26928.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578887|emb|CAC26929.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578889|emb|CAC26930.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578891|emb|CAC26931.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
gi|12578897|emb|CAC26934.1| ferulate-5-hydroxylase
[Arabidopsis thaliana] gi|12578899|emb|CAC26935.1|
ferulate-5-hydroxylase [Arabidopsis thaliana]
Length = 520
Score = 105 bits (262), Expect = 3e-22
Identities = 49/97 (50%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Frame = -3
Query: 528 WSDNAEEFYPERFTNNNF-DYRGHDFQFIPFGFGRRGCPGMNLGLASVKLVVAQLVHCFC 352
W+D + F P RF D++G +F+FIPFG GRR CPGM LGL ++ L VA ++HCF
Sbjct: 422 WTD-PDTFRPSRFLEPGVPDFKGSNFEFIPFGSGRRSCPGMQLGLYALDLAVAHILHCFT 480
Query: 351 WELPSNTTPNELDMSEKSGFSIPRAKHLFAVPTYRLL 241
W+LP P+ELDM++ G + P+A LFAVPT RL+
Sbjct: 481 WKLPDGMKPSELDMNDVFGLTAPKATRLFAVPTTRLI 517
>gb|AAG14961.1|AF214007_1 cytochrome p450-dependent monooxygenase [Brassica napus]
Length = 520
Score = 104 bits (260), Expect = 6e-22
Identities = 49/97 (50%), Positives = 66/97 (67%), Gaps = 1/97 (1%)
Frame = -3
Query: 528 WSDNAEEFYPERFTNNNF-DYRGHDFQFIPFGFGRRGCPGMNLGLASVKLVVAQLVHCFC 352
W D E F P RF D++G +F+FIPFG GRR CPGM LGL +++L VA ++HCF
Sbjct: 422 WVD-PETFRPSRFLEPGVPDFKGSNFEFIPFGSGRRSCPGMQLGLYALELAVAHILHCFT 480
Query: 351 WELPSNTTPNELDMSEKSGFSIPRAKHLFAVPTYRLL 241
W+LP P+ELDMS+ G + P+A L+AVP+ RL+
Sbjct: 481 WKLPDGMKPSELDMSDVFGLTAPKATRLYAVPSTRLI 517
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 398,391,563
Number of Sequences: 1393205
Number of extensions: 7860196
Number of successful extensions: 17869
Number of sequences better than 10.0: 1895
Number of HSP's better than 10.0 without gapping: 17251
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 17613
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)