Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004122A_C01 KMC004122A_c01
(888 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
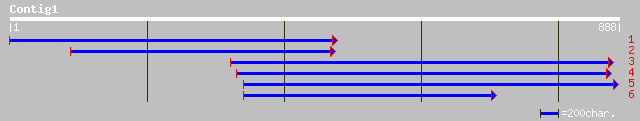
ref|NP_191099.1| putative protein; protein id: At3g55390.1, supp... 173 4e-42
gb|AAF68111.1|AC010793_6 F20B17.20 [Arabidopsis thaliana] 62 9e-09
ref|NP_198846.1| putative protein; protein id: At5g40300.1 [Arab... 53 5e-06
gb|AAO22748.1| unknown protein [Arabidopsis thaliana] 53 5e-06
ref|NP_567497.1| Expressed protein; protein id: At4g16442.1, sup... 50 3e-05
>ref|NP_191099.1| putative protein; protein id: At3g55390.1, supported by cDNA: 6434.
[Arabidopsis thaliana] gi|11358199|pir||T47678
hypothetical protein T22E16.50 - Arabidopsis thaliana
gi|7076782|emb|CAB75897.1| putative protein [Arabidopsis
thaliana] gi|21594014|gb|AAM65932.1| unknown
[Arabidopsis thaliana]
Length = 194
Score = 173 bits (438), Expect = 4e-42
Identities = 78/122 (63%), Positives = 97/122 (78%)
Frame = -3
Query: 886 DTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASA 707
D FRFV ANAIVA+YS+FEM VWE SR T++PE QVWFDFGHDQVF+YLLL+A +
Sbjct: 71 DAFRFVFVANAIVALYSVFEMGTCVWEFSRETTLWPEAFQVWFDFGHDQVFSYLLLSAGS 130
Query: 706 AGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVCFIINGSRF 527
A A+ RT++ DTCTA+ AFC+QSD+A+ LG+A F+FL F+S +GFRV CF+I GSRF
Sbjct: 131 AAAALARTMRGGDTCTANKAFCLQSDVAIGLGFAAFLFLAFSSCFSGFRVACFLITGSRF 190
Query: 526 HL 521
HL
Sbjct: 191 HL 192
>gb|AAF68111.1|AC010793_6 F20B17.20 [Arabidopsis thaliana]
Length = 393
Score = 62.4 bits (150), Expect = 9e-09
Identities = 31/112 (27%), Positives = 59/112 (52%), Gaps = 7/112 (6%)
Frame = -3
Query: 883 TFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAA 704
++++V+ A A +YSL ++ V+ + G+ + P Q W F DQ+F YL+++A +A
Sbjct: 275 SYQYVVGACAGTVLYSLLQLCLGVYRLVTGSPITPSRFQAWLCFTSDQLFCYLMMSAGSA 334
Query: 703 GTAM-------VRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLT 569
G+ + +R D C ++FC +++ L + F+FL +S T
Sbjct: 335 GSGVTNLNKTGIRHTPLPDFCKTLSSFCNHVALSLLLVFLSFIFLASSSFFT 386
>ref|NP_198846.1| putative protein; protein id: At5g40300.1 [Arabidopsis thaliana]
gi|10178139|dbj|BAB11584.1| gene_id:MPO12.1~unknown
protein [Arabidopsis thaliana]
Length = 270
Score = 53.1 bits (126), Expect = 5e-06
Identities = 39/106 (36%), Positives = 58/106 (53%)
Frame = -3
Query: 880 FRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG 701
FRF LAAN I VYS F + V+ +S L+ + +FG DQ+ AYLL ASA+
Sbjct: 160 FRFCLAANVIGFVYSGFMICDLVYLLSTSIRRSRHNLRHFLEFGLDQMLAYLL--ASAST 217
Query: 700 TAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 563
+A +R + + + ++ F + +VAL Y FV F SL +G+
Sbjct: 218 SASIR-VDDWQSNWGADKFPDLARASVALSYVSFVAFAFCSLASGY 262
>gb|AAO22748.1| unknown protein [Arabidopsis thaliana]
Length = 283
Score = 53.1 bits (126), Expect = 5e-06
Identities = 30/108 (27%), Positives = 57/108 (52%)
Frame = -3
Query: 880 FRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG 701
+RF L+ N + VYS F+ + + + + L+ F+F DQV AYLL++AS
Sbjct: 173 YRFCLSVNVVAFVYSSFQACDLAYHLVKEKHLISHHLRPLFEFIIDQVLAYLLMSAS--- 229
Query: 700 TAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 557
TA V + + + + F + ++A+ + F+ F+SL++G+ +
Sbjct: 230 TAAVTRVDDWVSNWGKDEFTEMASASIAMSFLAFLAFAFSSLISGYNL 277
>ref|NP_567497.1| Expressed protein; protein id: At4g16442.1, supported by cDNA:
7632. [Arabidopsis thaliana] gi|21595041|gb|AAM66067.1|
unknown [Arabidopsis thaliana]
gi|28393080|gb|AAO41974.1| unknown protein [Arabidopsis
thaliana]
Length = 182
Score = 50.4 bits (119), Expect = 3e-05
Identities = 36/111 (32%), Positives = 50/111 (44%), Gaps = 7/111 (6%)
Frame = -3
Query: 874 FVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTA 695
F++ AN I AVYSL + V V +G +F + L W F DQ AYL +AA AA
Sbjct: 56 FLVVANGIAAVYSLLQSVRCVVGTMKGKVLFSKPL-AWAFFSGDQAMAYLNVAAIAATAE 114
Query: 694 MVRTLKETD-------TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 563
+E + CT FC Q I V+ + + F S ++ F
Sbjct: 115 SGVIAREGEEDLQWMRVCTMYGKFCNQMAIGVSSALLASIAMVFVSCISAF 165
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 754,610,187
Number of Sequences: 1393205
Number of extensions: 17194868
Number of successful extensions: 109983
Number of sequences better than 10.0: 355
Number of HSP's better than 10.0 without gapping: 75478
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 100465
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 48218255001
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)