Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004103A_C01 KMC004103A_c01
(1059 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
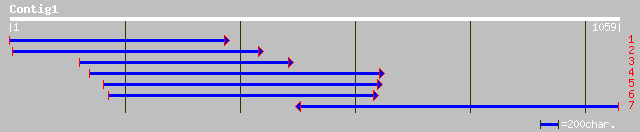
gb|AAF31706.1|AF221857_1 unknown [Euphorbia esula] 382 e-105
ref|NP_197671.1| putative protein; protein id: At5g22790.1, supp... 352 4e-96
pir||A84798 hypothetical protein At2g37860 [imported] - Arabidop... 343 2e-93
dbj|BAC16455.1| hypothetical protein~similar to Arabidopsis thal... 316 4e-85
ref|NP_181322.2| unknown protein; protein id: At2g37860.1, suppo... 210 3e-53
>gb|AAF31706.1|AF221857_1 unknown [Euphorbia esula]
Length = 268
Score = 382 bits (980), Expect = e-105
Identities = 184/214 (85%), Positives = 200/214 (92%)
Frame = -3
Query: 1057 KVGSEIVIDSCCATFAEVQKRGKDFWAEFELYAADLLVGVAVDIALVGLLAPYARIGKPS 878
KVG+EIVIDSCCATFAEVQKRGKDFW+EFELYAADLLVG+ VDIALVG+LAPYARIG+PS
Sbjct: 55 KVGTEIVIDSCCATFAEVQKRGKDFWSEFELYAADLLVGIVVDIALVGMLAPYARIGQPS 114
Query: 877 VSKGLLGRFQHACAALPSSVFEAQRPGCRFSVKQRIATYFYKGALYGSVGFGCGIIGQGI 698
+S+GLLG Q AC+ALPSSVFEA+RPGCRFS+KQR+ATYFYKG LYGSVGFGCG+IGQGI
Sbjct: 115 ISRGLLGNIQQACSALPSSVFEAERPGCRFSLKQRVATYFYKGVLYGSVGFGCGLIGQGI 174
Query: 697 ANAIMNAKRSISKSEEDIPIPPLLQSAALWGFFLAVSSNTRYQIINGLESLVEASPVAKK 518
AN IM AKRSI KSEEDIP+PPL+QSA LWG FLAVSSNTRYQIINGLESLVE SP+AKK
Sbjct: 175 ANLIMTAKRSIKKSEEDIPVPPLVQSAVLWGVFLAVSSNTRYQIINGLESLVEKSPLAKK 234
Query: 517 VPLVAMAFTVGVRFGNNIYGGMQFVDWAKWSGVQ 416
VP VAMAFTVGVRF NNIYGGMQF+DWAK SGVQ
Sbjct: 235 VPPVAMAFTVGVRFANNIYGGMQFIDWAKLSGVQ 268
>ref|NP_197671.1| putative protein; protein id: At5g22790.1, supported by cDNA:
gi_13605898, supported by cDNA: gi_19548024 [Arabidopsis
thaliana] gi|9758754|dbj|BAB09278.1|
gb|AAF31706.1~gene_id:K8E10.2~similar to unknown protein
[Arabidopsis thaliana]
gi|13605899|gb|AAK32935.1|AF367348_1 AT5g22790/K8E10_2
[Arabidopsis thaliana] gi|19548025|gb|AAL87376.1|
AT5g22790/K8E10_2 [Arabidopsis thaliana]
Length = 433
Score = 352 bits (904), Expect = 4e-96
Identities = 170/215 (79%), Positives = 191/215 (88%), Gaps = 1/215 (0%)
Frame = -3
Query: 1057 KVGSEIVIDSCCATFAEVQKRGKDFWAEFELYAADLLVGVAVDIALVGLLAPYARIGKPS 878
KVG+E+ IDSCCATFAEVQKRG+DFW+EFELYAADLLVG+ VD+ALVGLLAPYARIGKPS
Sbjct: 219 KVGTEVAIDSCCATFAEVQKRGEDFWSEFELYAADLLVGLVVDVALVGLLAPYARIGKPS 278
Query: 877 V-SKGLLGRFQHACAALPSSVFEAQRPGCRFSVKQRIATYFYKGALYGSVGFGCGIIGQG 701
V S GL + ACA+LPSSVFEA+RPGC+FSV QRIAT+FYKG LYGSVGFGCG+IGQG
Sbjct: 279 VASTGLFKDLKRACASLPSSVFEAERPGCKFSVNQRIATFFYKGLLYGSVGFGCGLIGQG 338
Query: 700 IANAIMNAKRSISKSEEDIPIPPLLQSAALWGFFLAVSSNTRYQIINGLESLVEASPVAK 521
IAN IM AKRS+ KSEED+PIPPL +SAALWG FL +SSN RYQIINGLE +VE S AK
Sbjct: 339 IANLIMTAKRSVKKSEEDVPIPPLFESAALWGVFLGLSSNARYQIINGLERVVEGSTAAK 398
Query: 520 KVPLVAMAFTVGVRFGNNIYGGMQFVDWAKWSGVQ 416
++P+VAMAFTVGVRF NN+YGGMQFVDWAK SGVQ
Sbjct: 399 RIPVVAMAFTVGVRFANNVYGGMQFVDWAKLSGVQ 433
>pir||A84798 hypothetical protein At2g37860 [imported] - Arabidopsis thaliana
Length = 432
Score = 343 bits (881), Expect = 2e-93
Identities = 166/214 (77%), Positives = 186/214 (86%)
Frame = -3
Query: 1057 KVGSEIVIDSCCATFAEVQKRGKDFWAEFELYAADLLVGVAVDIALVGLLAPYARIGKPS 878
K+G+EIVIDSCCAT AEVQKRGKDFWAEFELY ADLLVG V+IALVG+LAPY R G+PS
Sbjct: 219 KIGAEIVIDSCCATVAEVQKRGKDFWAEFELYVADLLVGTVVNIALVGMLAPYVRFGQPS 278
Query: 877 VSKGLLGRFQHACAALPSSVFEAQRPGCRFSVKQRIATYFYKGALYGSVGFGCGIIGQGI 698
S G LGR A ALPSSVFEA+RPGCRFS +QR+ATYFYKG +YG+VGFGCGI+GQGI
Sbjct: 279 ASPGFLGRMVFAYNALPSSVFEAERPGCRFSAQQRLATYFYKGIMYGAVGFGCGIVGQGI 338
Query: 697 ANAIMNAKRSISKSEEDIPIPPLLQSAALWGFFLAVSSNTRYQIINGLESLVEASPVAKK 518
AN IM AKR+I+KSEE+IP+PPL++SAALWG FL+VSSNTRYQIINGLE +VEASP AKK
Sbjct: 339 ANLIMTAKRNINKSEENIPVPPLIKSAALWGVFLSVSSNTRYQIINGLERVVEASPFAKK 398
Query: 517 VPLVAMAFTVGVRFGNNIYGGMQFVDWAKWSGVQ 416
P AMAFTVGVR NNIYGGMQFVDWA+ SG Q
Sbjct: 399 FPPAAMAFTVGVRLANNIYGGMQFVDWARLSGCQ 432
>dbj|BAC16455.1| hypothetical protein~similar to Arabidopsis thaliana chromosome5,
At5g22790 [Oryza sativa (japonica cultivar-group)]
Length = 438
Score = 316 bits (809), Expect = 4e-85
Identities = 154/214 (71%), Positives = 187/214 (86%)
Frame = -3
Query: 1057 KVGSEIVIDSCCATFAEVQKRGKDFWAEFELYAADLLVGVAVDIALVGLLAPYARIGKPS 878
K+G+EIVID+CCATFAEVQKRG++FW+EFELYAAD+LVGV V++ALVG+LAPYAR G S
Sbjct: 228 KIGTEIVIDTCCATFAEVQKRGEEFWSEFELYAADMLVGVVVNVALVGMLAPYARFGGGS 287
Query: 877 VSKGLLGRFQHACAALPSSVFEAQRPGCRFSVKQRIATYFYKGALYGSVGFGCGIIGQGI 698
S GLLGR +HA +LP SVFEA+RPG FS++QRI TYF+KG LYG+VGF CG++GQGI
Sbjct: 288 ASPGLLGRVRHAYDSLP-SVFEAERPGYSFSIQQRIGTYFFKGILYGTVGFFCGLVGQGI 346
Query: 697 ANAIMNAKRSISKSEEDIPIPPLLQSAALWGFFLAVSSNTRYQIINGLESLVEASPVAKK 518
AN IM AK S+ KS++D+P+PPLL+++ALW FL VSSNTRYQIINGLE +VEASPVAK+
Sbjct: 347 ANLIMTAK-SVKKSDDDVPVPPLLKTSALWA-FLGVSSNTRYQIINGLERVVEASPVAKR 404
Query: 517 VPLVAMAFTVGVRFGNNIYGGMQFVDWAKWSGVQ 416
VP V++AFTVGVRF NNIYGGMQFVDWA+ +G Q
Sbjct: 405 VPAVSLAFTVGVRFANNIYGGMQFVDWARMTGCQ 438
>ref|NP_181322.2| unknown protein; protein id: At2g37860.1, supported by cDNA:
gi_19698996 [Arabidopsis thaliana]
gi|19698997|gb|AAL91234.1| unknown protein [Arabidopsis
thaliana] gi|25084059|gb|AAN72164.1| unknown protein
[Arabidopsis thaliana]
Length = 347
Score = 210 bits (535), Expect = 3e-53
Identities = 100/129 (77%), Positives = 111/129 (85%)
Frame = -3
Query: 1057 KVGSEIVIDSCCATFAEVQKRGKDFWAEFELYAADLLVGVAVDIALVGLLAPYARIGKPS 878
K+G+EIVIDSCCAT AEVQKRGKDFWAEFELY ADLLVG V+IALVG+LAPY R G+PS
Sbjct: 219 KIGAEIVIDSCCATVAEVQKRGKDFWAEFELYVADLLVGTVVNIALVGMLAPYVRFGQPS 278
Query: 877 VSKGLLGRFQHACAALPSSVFEAQRPGCRFSVKQRIATYFYKGALYGSVGFGCGIIGQGI 698
S G LGR A ALPSSVFEA+RPGCRFS +QR+ATYFYKG +YG+VGFGCGI+GQGI
Sbjct: 279 ASPGFLGRMVFAYNALPSSVFEAERPGCRFSAQQRLATYFYKGIMYGAVGFGCGIVGQGI 338
Query: 697 ANAIMNAKR 671
AN IM AKR
Sbjct: 339 ANLIMTAKR 347
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 951,719,953
Number of Sequences: 1393205
Number of extensions: 22051190
Number of successful extensions: 69949
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 62152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 69039
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 62912456556
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)