Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004085A_C01 KMC004085A_c01
(550 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
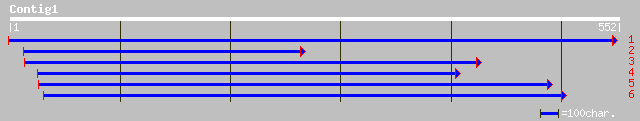
emb|CAB50925.1| translocon Tic40 [Pisum sativum] 112 3e-24
gb|AAN75219.1| chloroplast protein translocon component Tic40 pr... 112 3e-24
ref|NP_197165.1| translocon Tic40-like protein; protein id: At5g... 103 1e-21
ref|NP_112384.1| suppression of tumorigenicity 13 (colon carcino... 54 9e-07
ref|XP_145369.1| similar to RIKEN cDNA 3110002K08 [Mus musculus] 54 9e-07
>emb|CAB50925.1| translocon Tic40 [Pisum sativum]
Length = 436
Score = 112 bits (280), Expect = 3e-24
Identities = 54/59 (91%), Positives = 57/59 (96%)
Frame = -3
Query: 548 SKIMAHPDVAMAFQNPRVQAAIMDCSQNPLNIAKYQNDKEVMDVFNKISELFPGVSG*P 372
SKIMA+PDVAMAFQNPRVQAAIMDCSQNP++I KYQNDKEVMDVFNKISELFPGVSG P
Sbjct: 378 SKIMANPDVAMAFQNPRVQAAIMDCSQNPMSIVKYQNDKEVMDVFNKISELFPGVSGPP 436
>gb|AAN75219.1| chloroplast protein translocon component Tic40 precursor [Pisum
sativum]
Length = 436
Score = 112 bits (280), Expect = 3e-24
Identities = 54/59 (91%), Positives = 57/59 (96%)
Frame = -3
Query: 548 SKIMAHPDVAMAFQNPRVQAAIMDCSQNPLNIAKYQNDKEVMDVFNKISELFPGVSG*P 372
SKIMA+PDVAMAFQNPRVQAAIMDCSQNP++I KYQNDKEVMDVFNKISELFPGVSG P
Sbjct: 378 SKIMANPDVAMAFQNPRVQAAIMDCSQNPMSIVKYQNDKEVMDVFNKISELFPGVSGPP 436
>ref|NP_197165.1| translocon Tic40-like protein; protein id: At5g16620.1, supported
by cDNA: gi_16226312, supported by cDNA: gi_20260221
[Arabidopsis thaliana] gi|10176971|dbj|BAB10189.1|
translocon Tic40-like protein [Arabidopsis thaliana]
gi|16226313|gb|AAL16131.1|AF428299_1 AT5g16620/MTG13_6
[Arabidopsis thaliana] gi|20260222|gb|AAM13009.1|
translocon Tic40-like protein [Arabidopsis thaliana]
Length = 447
Score = 103 bits (258), Expect = 1e-21
Identities = 47/57 (82%), Positives = 55/57 (96%)
Frame = -3
Query: 548 SKIMAHPDVAMAFQNPRVQAAIMDCSQNPLNIAKYQNDKEVMDVFNKISELFPGVSG 378
SKIM +PDVAMAFQNPRVQAA+M+CS+NP+NI KYQNDKEVMDVFNKIS+LFPG++G
Sbjct: 391 SKIMENPDVAMAFQNPRVQAALMECSENPMNIMKYQNDKEVMDVFNKISQLFPGMTG 447
>ref|NP_112384.1| suppression of tumorigenicity 13 (colon carcinoma) Hsp70-interac
[Rattus norvegicus] gi|1708200|sp|P50503|HIP_RAT
HSC70-INTERACTING PROTEIN gi|4379408|emb|CAA57546.1|
Hsc70-interacting protein [Rattus norvegicus]
Length = 368
Score = 54.3 bits (129), Expect = 9e-07
Identities = 24/56 (42%), Positives = 39/56 (68%)
Frame = -3
Query: 548 SKIMAHPDVAMAFQNPRVQAAIMDCSQNPLNIAKYQNDKEVMDVFNKISELFPGVS 381
++I++ P+V A Q+P V A D +QNP N++KYQN+ +VM++ +K+S F G S
Sbjct: 313 NEILSDPEVLAAMQDPEVMVAFQDVAQNPSNMSKYQNNPKVMNLISKLSAKFGGHS 368
>ref|XP_145369.1| similar to RIKEN cDNA 3110002K08 [Mus musculus]
Length = 271
Score = 54.3 bits (129), Expect = 9e-07
Identities = 23/56 (41%), Positives = 40/56 (71%)
Frame = -3
Query: 548 SKIMAHPDVAMAFQNPRVQAAIMDCSQNPLNIAKYQNDKEVMDVFNKISELFPGVS 381
S+I+++P+V + QNP V A D +QNP N++KYQ++ +V+++ +K+S F G S
Sbjct: 211 SEILSNPEVLASMQNPEVMVAFQDVAQNPSNLSKYQSNPKVVNLISKLSAKFGGQS 266
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,050,007
Number of Sequences: 1393205
Number of extensions: 9423700
Number of successful extensions: 20103
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 19615
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20102
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)