Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
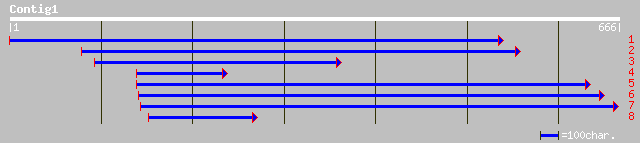
Query= KMC004062A_C01 KMC004062A_c01
(663 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL67496.1|AF459411_1 senescence-associated putative protein ... 109 4e-23
ref|NP_566492.1| expressed protein; protein id: At3g14750.1, sup... 94 1e-18
dbj|BAC06972.1| similar to senescence-associated protein [Oryza ... 92 5e-18
dbj|BAB02406.1| gb|AAD10662.1~gene_id:MIE1.26~similar to unknown... 83 3e-15
gb|AAO60017.1| hypothetical protein [Oryza sativa (japonica cult... 57 2e-07
>gb|AAL67496.1|AF459411_1 senescence-associated putative protein [Narcissus pseudonarcissus]
Length = 128
Score = 109 bits (272), Expect = 4e-23
Identities = 61/108 (56%), Positives = 70/108 (64%), Gaps = 8/108 (7%)
Frame = -3
Query: 658 GQVMEKKLISMAREMEKLRAEISNAEKRARAAAAAGNPG---PGYNANYGNAETGYAGNP 488
GQVMEK L+SMARE+EKLRAE++NAEKRARAAAAAG+ G Y NY N + Y GNP
Sbjct: 21 GQVMEKNLVSMAREVEKLRAEVANAEKRARAAAAAGSQGFAAATYGMNYANPDPNYGGNP 80
Query: 487 YP-AYYGMNPVQPG--AENFPQYGPGPGPGPG--AWGAYDMQRAQGHR 359
YP A Y N PG A+ PQY G G G +WG+YDMQ R
Sbjct: 81 YPAASYTTNAQVPGNAADGGPQYAAGAGSAAGHASWGSYDMQHTHARR 128
>ref|NP_566492.1| expressed protein; protein id: At3g14750.1, supported by cDNA:
gi_14334819, supported by cDNA: gi_15293200 [Arabidopsis
thaliana] gi|14334820|gb|AAK59588.1| unknown protein
[Arabidopsis thaliana] gi|15293201|gb|AAK93711.1|
unknown protein [Arabidopsis thaliana]
Length = 331
Score = 94.4 bits (233), Expect = 1e-18
Identities = 52/102 (50%), Positives = 61/102 (58%), Gaps = 4/102 (3%)
Frame = -3
Query: 661 HGQVMEKKLISMAREMEKLRAEISNAEKRARAAAAAGNP-GPGYNANYGNAETGYAGNPY 485
HG++ME KL++MARE+EKLRAEI+N+E A A GNP G Y YGN E GY NPY
Sbjct: 223 HGKIMEHKLVAMARELEKLRAEIANSETSAYANGPVGNPGGVAYGGGYGNPEAGYPVNPY 282
Query: 484 PAYYGMNPVQPGAENF--PQYGPGPGPGPGAW-GAYDMQRAQ 368
Y MNP Q G + P YGP AW G YD Q+ Q
Sbjct: 283 QPNYTMNPAQTGVVGYYPPPYGP-----QAAWAGGYDPQQQQ 319
>dbj|BAC06972.1| similar to senescence-associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 185
Score = 92.4 bits (228), Expect = 5e-18
Identities = 55/103 (53%), Positives = 65/103 (62%), Gaps = 3/103 (2%)
Frame = -3
Query: 658 GQVMEKKLISMAREMEKLRAEISNAEKRARAAAAAGNPGPGYNANYGNAETGYAGNPYPA 479
GQ M+KKLIS+A E+EKLRAE AEKR+RAA + GN Y YGN + YA NPY A
Sbjct: 90 GQEMQKKLISVASEVEKLRAE---AEKRSRAAVSGGNQV--YVGGYGNPKAAYAANPYNA 144
Query: 478 YYGMNPVQP---GAENFPQYGPGPGPGPGAWGAYDMQRAQGHR 359
Y +N P A++ Q+GPG P WGAYDMQRA G R
Sbjct: 145 GYNINQPHPQANTADSGSQFGPGSTHAP--WGAYDMQRATGRR 185
>dbj|BAB02406.1| gb|AAD10662.1~gene_id:MIE1.26~similar to unknown protein
[Arabidopsis thaliana]
Length = 349
Score = 83.2 bits (204), Expect = 3e-15
Identities = 52/120 (43%), Positives = 61/120 (50%), Gaps = 22/120 (18%)
Frame = -3
Query: 661 HGQVMEKKLISMAREMEKLRAEISNAEKRARAAAAAGNP-GPGYNANYGNAETGYAGNPY 485
HG++ME KL++MARE+EKLRAEI+N+E A A GNP G Y YGN E GY NPY
Sbjct: 223 HGKIMEHKLVAMARELEKLRAEIANSETSAYANGPVGNPGGVAYGGGYGNPEAGYPVNPY 282
Query: 484 PAYYGMNP------------------VQPGAENF--PQYGPGPGPGPGAW-GAYDMQRAQ 368
Y MNP Q G + P YGP AW G YD Q+ Q
Sbjct: 283 QPNYTMNPTNLSQQCELMSMKPVTEYAQTGVVGYYPPPYGP-----QAAWAGGYDPQQQQ 337
>gb|AAO60017.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 345
Score = 57.0 bits (136), Expect = 2e-07
Identities = 31/63 (49%), Positives = 41/63 (64%), Gaps = 1/63 (1%)
Frame = -3
Query: 655 QVMEKKLISMAREMEKLRAEISNAEKRARA-AAAAGNPGPGYNANYGNAETGYAGNPYPA 479
+ MEK +I++A E+EKLR +++NAEKRA A A A PG+ YGN+E Y P PA
Sbjct: 214 KAMEKNMIAVASEIEKLRGDLANAEKRATAVTATAPVANPGFPTTYGNSEATY---PAPA 270
Query: 478 YYG 470
YG
Sbjct: 271 AYG 273
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 600,498,437
Number of Sequences: 1393205
Number of extensions: 14809719
Number of successful extensions: 74110
Number of sequences better than 10.0: 346
Number of HSP's better than 10.0 without gapping: 59939
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 71719
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28572683052
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)