Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004057A_C01 KMC004057A_c01
(572 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
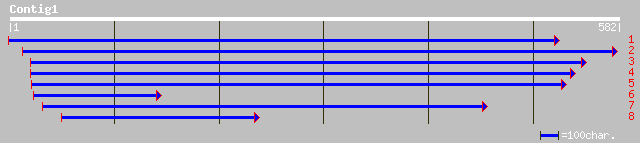
ref|NP_567928.1| carbonate dehydratase - like protein; protein i... 188 4e-47
pir||T04975 carbonate dehydratase homolog T16L1.70 - Arabidopsis... 188 4e-47
gb|AAG50705.1|AC079604_12 carbonic anhydrase, putative [Arabidop... 147 7e-35
ref|NP_176114.1| carbonic anhydrase, putative; protein id: At1g5... 143 1e-33
pir||T01481 carbonate dehydratase homolog F17O7.5 - Arabidopsis ... 139 3e-32
>ref|NP_567928.1| carbonate dehydratase - like protein; protein id: At4g33580.1,
supported by cDNA: 6589., supported by cDNA: gi_14334477
[Arabidopsis thaliana] gi|14334478|gb|AAK59437.1|
putative carbonate dehydratase [Arabidopsis thaliana]
gi|21594039|gb|AAM65957.1| carbonate dehydratase-like
protein [Arabidopsis thaliana]
gi|21689751|gb|AAM67519.1| putative carbonate
dehydratase [Arabidopsis thaliana]
Length = 301
Score = 188 bits (478), Expect = 4e-47
Identities = 91/142 (64%), Positives = 110/142 (77%), Gaps = 4/142 (2%)
Frame = -1
Query: 572 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDA-SASFIKNWVVIGKNARIKTEAAA 396
AALEF+VNTL +ENI VIGHS CGGI+ALM M+D+ S SFI NWVV+GK A+ T+A A
Sbjct: 160 AALEFSVNTLNVENILVIGHSRCGGIQALMKMEDEGDSRSFIHNWVVVGKKAKESTKAVA 219
Query: 395 SNLSFDEQCSHCEKESINHSLLNLLTYPWIEQKVANGELMIHGGYYNFIDCSFEKWTLDY 216
SNL FD QC HCEK SINHSL LL YPWIE+KV G L +HGGYYNF+DC+FEKWT+DY
Sbjct: 220 SNLHFDHQCQHCEKASINHSLERLLGYPWIEEKVRQGSLSLHGGYYNFVDCTFEKWTVDY 279
Query: 215 ---RGTEVDGNGRIATQHQAFW 159
RG + +G+G IA + ++ W
Sbjct: 280 AASRGKKKEGSG-IAVKDRSVW 300
>pir||T04975 carbonate dehydratase homolog T16L1.70 - Arabidopsis thaliana
gi|3549660|emb|CAA20571.1| carbonate dehydratase-like
protein [Arabidopsis thaliana]
gi|7270306|emb|CAB80075.1| carbonate dehydratase-like
protein [Arabidopsis thaliana]
Length = 173
Score = 188 bits (478), Expect = 4e-47
Identities = 91/142 (64%), Positives = 110/142 (77%), Gaps = 4/142 (2%)
Frame = -1
Query: 572 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDA-SASFIKNWVVIGKNARIKTEAAA 396
AALEF+VNTL +ENI VIGHS CGGI+ALM M+D+ S SFI NWVV+GK A+ T+A A
Sbjct: 32 AALEFSVNTLNVENILVIGHSRCGGIQALMKMEDEGDSRSFIHNWVVVGKKAKESTKAVA 91
Query: 395 SNLSFDEQCSHCEKESINHSLLNLLTYPWIEQKVANGELMIHGGYYNFIDCSFEKWTLDY 216
SNL FD QC HCEK SINHSL LL YPWIE+KV G L +HGGYYNF+DC+FEKWT+DY
Sbjct: 92 SNLHFDHQCQHCEKASINHSLERLLGYPWIEEKVRQGSLSLHGGYYNFVDCTFEKWTVDY 151
Query: 215 ---RGTEVDGNGRIATQHQAFW 159
RG + +G+G IA + ++ W
Sbjct: 152 AASRGKKKEGSG-IAVKDRSVW 172
>gb|AAG50705.1|AC079604_12 carbonic anhydrase, putative [Arabidopsis thaliana]
Length = 290
Score = 147 bits (372), Expect = 7e-35
Identities = 70/119 (58%), Positives = 90/119 (74%), Gaps = 2/119 (1%)
Frame = -1
Query: 572 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDAS--ASFIKNWVVIGKNARIKTEAA 399
+ALEFAV TLQ+ENI V+GHS CGGI ALMS Q+ +S ++ WV+ GK A+++T+ A
Sbjct: 154 SALEFAVTTLQVENIIVMGHSNCGGIAALMSHQNHQGQHSSLVERWVMNGKAAKLRTQLA 213
Query: 398 ASNLSFDEQCSHCEKESINHSLLNLLTYPWIEQKVANGELMIHGGYYNFIDCSFEKWTL 222
+S+LSFDEQC +CEKESI S++NL+TY WI +V GE+ IHG YYN DCS EKW L
Sbjct: 214 SSHLSFDEQCRNCEKESIKDSVMNLITYSWIRDRVKRGEVKIHGCYYNLSDCSLEKWRL 272
>ref|NP_176114.1| carbonic anhydrase, putative; protein id: At1g58180.1 [Arabidopsis
thaliana] gi|25404198|pir||B96615 probable carbonic
anhydrase T18I24.9 [imported] - Arabidopsis thaliana
gi|12321393|gb|AAG50771.1|AC079131_16 carbonic
anhydrase, putative [Arabidopsis thaliana]
Length = 286
Score = 143 bits (361), Expect = 1e-33
Identities = 69/117 (58%), Positives = 86/117 (72%)
Frame = -1
Query: 572 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDASASFIKNWVVIGKNARIKTEAAAS 393
+ALEFAV TLQ+ENI V+GHS CGGI ALMS Q+ WV+ GK A+++T+ A+S
Sbjct: 154 SALEFAVTTLQVENIIVMGHSNCGGIAALMSHQNHQGQH--SRWVMNGKAAKLRTQLASS 211
Query: 392 NLSFDEQCSHCEKESINHSLLNLLTYPWIEQKVANGELMIHGGYYNFIDCSFEKWTL 222
+LSFDEQC +CEKESI S++NL+TY WI +V GE+ IHG YYN DCS EKW L
Sbjct: 212 HLSFDEQCRNCEKESIKDSVMNLITYSWIRDRVKRGEVKIHGCYYNLSDCSLEKWRL 268
>pir||T01481 carbonate dehydratase homolog F17O7.5 - Arabidopsis thaliana
gi|3176676|gb|AAC18799.1| Similar to carbonic anhydrase
gb|L19255 from Nicotiana tabacum. ESTs gb|AA597643,
gb|T45390, gb|T43963 and gb|AA597734 come from this
gene. [Arabidopsis thaliana] gi|21593413|gb|AAM65380.1|
carbonic anhydrase, putative [Arabidopsis thaliana]
Length = 258
Score = 139 bits (349), Expect = 3e-32
Identities = 65/125 (52%), Positives = 93/125 (74%), Gaps = 3/125 (2%)
Frame = -1
Query: 572 AALEFAVNTLQLENIFVIGHSCCGGIRALMSMQDDAS---ASFIKNWVVIGKNARIKTEA 402
AA+E+AV L++ENI VIGHSCCGGI+ LMS++DDA+ + FI+NWV IG +AR K +
Sbjct: 128 AAVEYAVVHLKVENILVIGHSCCGGIKGLMSIEDDAAPTQSDFIENWVKIGASARNKIKE 187
Query: 401 AASNLSFDEQCSHCEKESINHSLLNLLTYPWIEQKVANGELMIHGGYYNFIDCSFEKWTL 222
+LS+D+QC+ CEKE++N SL NLL+YP++ +V L I GG+YNF+ +F+ W L
Sbjct: 188 EHKDLSYDDQCNKCEKEAVNVSLGNLLSYPFVRAEVVKNTLAIRGGHYNFVKGTFDLWEL 247
Query: 221 DYRGT 207
D++ T
Sbjct: 248 DFKTT 252
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,757,453
Number of Sequences: 1393205
Number of extensions: 10637413
Number of successful extensions: 25253
Number of sequences better than 10.0: 169
Number of HSP's better than 10.0 without gapping: 24254
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25163
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)