Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
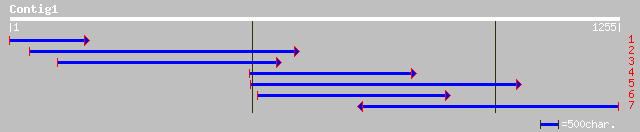
Query= KMC004056A_C01 KMC004056A_c01
(1242 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172478.1| tat-binding protein, putative; protein id: At1g... 276 7e-77
pir||T00626 branched-chain amino acid aminotransferase homolog T... 276 7e-77
gb|AAK57535.1| branched-chain amino acid aminotransferase [Capsi... 259 5e-71
gb|AAO00685.1| Unknown protein [Oryza sativa (japonica cultivar-... 249 3e-68
gb|AAK43507.1|AC020666_17 putative aminotransferase [Oryza sativ... 252 6e-67
>ref|NP_172478.1| tat-binding protein, putative; protein id: At1g10070.1, supported by
cDNA: gi_13877744, supported by cDNA: gi_15293208
[Arabidopsis thaliana] gi|26391680|sp|Q9M439|BCA2_ARATH
Branched-chain amino acid aminotransferase 2, chloroplast
precursor (Atbcat-2) gi|8249004|emb|CAB93128.1|
branched-chain amino acid transaminase [Arabidopsis
thaliana] gi|13877745|gb|AAK43950.1|AF370135_1 putative
tat-binding protein [Arabidopsis thaliana]
gi|15293209|gb|AAK93715.1| putative tat-binding protein
[Arabidopsis thaliana]
Length = 388
Score = 276 bits (706), Expect(2) = 7e-77
Identities = 136/188 (72%), Positives = 160/188 (84%)
Frame = -1
Query: 1242 MRPLLLGSGPVLCLAPAPEYTFLVYASPVRNYFKEGSAPLSLFVEEDFDRASRRGTGSVK 1063
+RPLL+GSGP+L L PAPEYTF+VYASPV NYFKEG A L+L+VEE++ RA+ G G VK
Sbjct: 176 IRPLLMGSGPILGLGPAPEYTFIVYASPVGNYFKEGMAALNLYVEEEYVRAAPGGAGGVK 235
Query: 1062 TISNYAPVLMAQLRAKNRGFSDVLYLDSETKKNLEEVSSCNIFIAKGKCISTPATNGTIL 883
+I+NYAPVL A RAK+RGFSDVLYLDS KK LEE SSCN+F+ KG+ ISTPATNGTIL
Sbjct: 236 SITNYAPVLKALSRAKSRGFSDVLYLDSVKKKYLEEASSCNVFVVKGRTISTPATNGTIL 295
Query: 882 AGITRKSVIEIAGDLGYQVEEHAVNVDELMEADEVFCTGTAVGVAPVGSITYQDRRAEYK 703
GITRKSV+EIA D GYQV E AV+VDE+M+ADEVFCTGTAV VAPVG+ITYQ++R EYK
Sbjct: 296 EGITRKSVMEIASDQGYQVVEKAVHVDEVMDADEVFCTGTAVVVAPVGTITYQEKRVEYK 355
Query: 702 TGSGTICQ 679
TG ++CQ
Sbjct: 356 TGDESVCQ 363
Score = 35.0 bits (79), Expect(2) = 7e-77
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = -2
Query: 704 KQDPEPFVNELYKTISGIQTGTIEDKKGWIVEID 603
K E +L + GIQTG IED KGW+ +I+
Sbjct: 355 KTGDESVCQKLRSVLVGIQTGLIEDNKGWVTDIN 388
>pir||T00626 branched-chain amino acid aminotransferase homolog T27I1.9 -
Arabidopsis thaliana gi|3540183|gb|AAC34333.1| Highly
Similar to branched-chain amino acid aminotransferase
[Arabidopsis thaliana]
Length = 318
Score = 276 bits (706), Expect(2) = 7e-77
Identities = 136/188 (72%), Positives = 160/188 (84%)
Frame = -1
Query: 1242 MRPLLLGSGPVLCLAPAPEYTFLVYASPVRNYFKEGSAPLSLFVEEDFDRASRRGTGSVK 1063
+RPLL+GSGP+L L PAPEYTF+VYASPV NYFKEG A L+L+VEE++ RA+ G G VK
Sbjct: 106 IRPLLMGSGPILGLGPAPEYTFIVYASPVGNYFKEGMAALNLYVEEEYVRAAPGGAGGVK 165
Query: 1062 TISNYAPVLMAQLRAKNRGFSDVLYLDSETKKNLEEVSSCNIFIAKGKCISTPATNGTIL 883
+I+NYAPVL A RAK+RGFSDVLYLDS KK LEE SSCN+F+ KG+ ISTPATNGTIL
Sbjct: 166 SITNYAPVLKALSRAKSRGFSDVLYLDSVKKKYLEEASSCNVFVVKGRTISTPATNGTIL 225
Query: 882 AGITRKSVIEIAGDLGYQVEEHAVNVDELMEADEVFCTGTAVGVAPVGSITYQDRRAEYK 703
GITRKSV+EIA D GYQV E AV+VDE+M+ADEVFCTGTAV VAPVG+ITYQ++R EYK
Sbjct: 226 EGITRKSVMEIASDQGYQVVEKAVHVDEVMDADEVFCTGTAVVVAPVGTITYQEKRVEYK 285
Query: 702 TGSGTICQ 679
TG ++CQ
Sbjct: 286 TGDESVCQ 293
Score = 35.0 bits (79), Expect(2) = 7e-77
Identities = 15/34 (44%), Positives = 20/34 (58%)
Frame = -2
Query: 704 KQDPEPFVNELYKTISGIQTGTIEDKKGWIVEID 603
K E +L + GIQTG IED KGW+ +I+
Sbjct: 285 KTGDESVCQKLRSVLVGIQTGLIEDNKGWVTDIN 318
>gb|AAK57535.1| branched-chain amino acid aminotransferase [Capsicum annuum]
Length = 385
Score = 259 bits (663), Expect(2) = 5e-71
Identities = 126/188 (67%), Positives = 151/188 (80%)
Frame = -1
Query: 1242 MRPLLLGSGPVLCLAPAPEYTFLVYASPVRNYFKEGSAPLSLFVEEDFDRASRRGTGSVK 1063
+RPLL+G+GP+L LAPAPEYTFLVYA PV NYFKEG+APL+L+VEED RASR G G VK
Sbjct: 166 IRPLLIGTGPILGLAPAPEYTFLVYACPVGNYFKEGTAPLNLYVEEDVHRASRGGAGGVK 225
Query: 1062 TISNYAPVLMAQLRAKNRGFSDVLYLDSETKKNLEEVSSCNIFIAKGKCISTPATNGTIL 883
+I+NYAPVL A +AK G+SDVLY+D+ KK +EEVSSCNIF+ KG +STP GTIL
Sbjct: 226 SITNYAPVLKAMKKAKANGYSDVLYVDAVNKKYIEEVSSCNIFVVKGNVVSTPIAKGTIL 285
Query: 882 AGITRKSVIEIAGDLGYQVEEHAVNVDELMEADEVFCTGTAVGVAPVGSITYQDRRAEYK 703
G+TRKS+++IA DLGY VEE + DEL+ ADEVFCTGTAVGVAPVGSITY+ +R EYK
Sbjct: 286 EGVTRKSIMDIALDLGYTVEERLIEADELISADEVFCTGTAVGVAPVGSITYKGQRIEYK 345
Query: 702 TGSGTICQ 679
S C+
Sbjct: 346 ISSDLSCK 353
Score = 32.0 bits (71), Expect(2) = 5e-71
Identities = 12/25 (48%), Positives = 18/25 (72%)
Frame = -2
Query: 677 ELYKTISGIQTGTIEDKKGWIVEID 603
+ Y + GIQ G I+D++ WIVEI+
Sbjct: 354 KFYSRLVGIQKGVIKDERNWIVEIE 378
>gb|AAO00685.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
gi|27356673|gb|AAO06962.1| Putative branched-chain amino
acid aminotransferase [Oryza sativa (japonica
cultivar-group)]
Length = 415
Score = 249 bits (635), Expect(2) = 3e-68
Identities = 124/188 (65%), Positives = 150/188 (78%)
Frame = -1
Query: 1242 MRPLLLGSGPVLCLAPAPEYTFLVYASPVRNYFKEGSAPLSLFVEEDFDRASRRGTGSVK 1063
+RPLL+GSGP+L LAPAPEYTFL+YA+PV YFKEG AP++L VE+ RA GTG VK
Sbjct: 203 IRPLLIGSGPILGLAPAPEYTFLIYAAPVGTYFKEGLAPINLVVEDSIHRAMPGGTGGVK 262
Query: 1062 TISNYAPVLMAQLRAKNRGFSDVLYLDSETKKNLEEVSSCNIFIAKGKCISTPATNGTIL 883
TI+NYAPVL AQ+ AK+RGF+DVLYLD+ K LEE SSCN+FI K ++TPAT GTIL
Sbjct: 263 TITNYAPVLKAQMDAKSRGFTDVLYLDAVHKTYLEEASSCNLFIVKDGVVATPATVGTIL 322
Query: 882 AGITRKSVIEIAGDLGYQVEEHAVNVDELMEADEVFCTGTAVGVAPVGSITYQDRRAEYK 703
GITRKSVIE+A D GYQVEE V++D+L+ ADEVFCTGTAV VAPV S+TY +R E++
Sbjct: 323 PGITRKSVIELARDRGYQVEERLVSIDDLVGADEVFCTGTAVVVAPVSSVTYHGQRYEFR 382
Query: 702 TGSGTICQ 679
TG T+ Q
Sbjct: 383 TGHDTLSQ 390
Score = 33.5 bits (75), Expect(2) = 3e-68
Identities = 14/24 (58%), Positives = 17/24 (70%)
Frame = -2
Query: 674 LYKTISGIQTGTIEDKKGWIVEID 603
L+ T++ IQ G EDKKGW V ID
Sbjct: 392 LHTTLTSIQMGLAEDKKGWTVAID 415
>gb|AAK43507.1|AC020666_17 putative aminotransferase [Oryza sativa (japonica cultivar-group)]
Length = 381
Score = 252 bits (643), Expect(2) = 6e-67
Identities = 127/184 (69%), Positives = 148/184 (80%)
Frame = -1
Query: 1242 MRPLLLGSGPVLCLAPAPEYTFLVYASPVRNYFKEGSAPLSLFVEEDFDRASRRGTGSVK 1063
+RPLL+GSG VL LAPAPEYTF+++ SPV NYFKEG AP++L VE+ F RA+ GTGSVK
Sbjct: 167 IRPLLMGSGAVLGLAPAPEYTFIIFVSPVGNYFKEGLAPINLIVEDKFHRATPGGTGSVK 226
Query: 1062 TISNYAPVLMAQLRAKNRGFSDVLYLDSETKKNLEEVSSCNIFIAKGKCISTPATNGTIL 883
TI NYA VLMAQ AK +G+SDVLYLD+ KK LEEVSSCNIF+ KG ISTPA GTIL
Sbjct: 227 TIGNYASVLMAQKIAKEKGYSDVLYLDAVHKKYLEEVSSCNIFVVKGNVISTPAVKGTIL 286
Query: 882 AGITRKSVIEIAGDLGYQVEEHAVNVDELMEADEVFCTGTAVGVAPVGSITYQDRRAEYK 703
GITRKS+I++A G+QVEE V+VDEL+EADEVFCTGTAV V+PVGSITYQ +R EY
Sbjct: 287 PGITRKSIIDVALSKGFQVEERLVSVDELLEADEVFCTGTAVVVSPVGSITYQGKRVEYA 346
Query: 702 TGSG 691
G
Sbjct: 347 GNKG 350
Score = 26.2 bits (56), Expect(2) = 6e-67
Identities = 9/24 (37%), Positives = 16/24 (66%)
Frame = -2
Query: 677 ELYKTISGIQTGTIEDKKGWIVEI 606
+LY +++ +Q G ED GW V++
Sbjct: 357 QLYTSLTSLQMGQAEDWLGWTVQL 380
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,129,606,689
Number of Sequences: 1393205
Number of extensions: 26453488
Number of successful extensions: 74059
Number of sequences better than 10.0: 302
Number of HSP's better than 10.0 without gapping: 68640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 73766
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 78392734679
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)