Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004053A_C01 KMC004053A_c01
(509 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
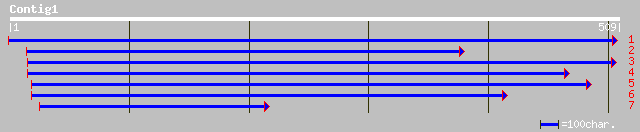
gb|AAG38521.1|AF283536_1 cystatin-like protein [Citrus x paradisi] 107 6e-23
ref|NP_199566.1| putative protein; protein id: At5g47550.1, supp... 104 5e-22
gb|AAM64661.1| cystatin-like protein [Arabidopsis thaliana] 104 7e-22
pir||T07822 cystein proteinase inhibitor - cucumber gi|3237190|d... 95 5e-19
ref|NP_193383.1| cysteine proteinase inhibitor like protein; pro... 86 3e-16
>gb|AAG38521.1|AF283536_1 cystatin-like protein [Citrus x paradisi]
Length = 118
Score = 107 bits (268), Expect = 6e-23
Identities = 56/117 (47%), Positives = 79/117 (66%)
Frame = -3
Query: 495 RVLIVSLLILSAVSGFSAALRQKGAFVGGYTPIKDLNDPHVKEIAHFAVSEYDKQSGAKL 316
R + +L LS V +A R KGA VGG+ PI+D + HV EI FAV+EY+KQS + L
Sbjct: 4 RFCCLIVLFLSVVPLLAAGDR-KGALVGGWKPIEDPKEKHVMEIGQFAVTEYNKQSKSAL 62
Query: 315 RLVKVVSGDSQVVAGTNYRLVIAAKGGSRSAAASNYEALVYERSWEHFRNLTSFKPV 145
+ V G++QVV+GTNYRL++ K G + +EA+V+E+ WEHF++LTSFKP+
Sbjct: 63 KFESVEKGETQVVSGTNYRLILVVKDG---PSTKKFEAVVWEKPWEHFKSLTSFKPM 116
>ref|NP_199566.1| putative protein; protein id: At5g47550.1, supported by cDNA:
31680., supported by cDNA: gi_18252838 [Arabidopsis
thaliana] gi|9758783|dbj|BAB09081.1|
gene_id:MNJ7.14~pir||H71431~similar to unknown protein
[Arabidopsis thaliana] gi|18252839|gb|AAL62346.1|
putative protein [Arabidopsis thaliana]
Length = 122
Score = 104 bits (260), Expect = 5e-22
Identities = 55/115 (47%), Positives = 76/115 (65%)
Frame = -3
Query: 486 IVSLLILSAVSGFSAALRQKGAFVGGYTPIKDLNDPHVKEIAHFAVSEYDKQSGAKLRLV 307
+V LL+LS V A VGG++PI ++ DP V EI FAVSEY+K+S + L+
Sbjct: 5 VVFLLLLSLVVVLLPLYASAAARVGGWSPISNVTDPQVVEIGEFAVSEYNKRSESGLKFE 64
Query: 306 KVVSGDSQVVAGTNYRLVIAAKGGSRSAAASNYEALVYERSWEHFRNLTSFKPVH 142
VVSG++QVV+GTNYRL +AA G + NY A+V+++ W FRNLTSF+P +
Sbjct: 65 TVVSGETQVVSGTNYRLKVAANDG--DGVSKNYLAIVWDKPWMKFRNLTSFEPAN 117
>gb|AAM64661.1| cystatin-like protein [Arabidopsis thaliana]
Length = 122
Score = 104 bits (259), Expect = 7e-22
Identities = 55/115 (47%), Positives = 76/115 (65%)
Frame = -3
Query: 486 IVSLLILSAVSGFSAALRQKGAFVGGYTPIKDLNDPHVKEIAHFAVSEYDKQSGAKLRLV 307
+V LL+LS V A VGG++PI ++ DP V EI FAVSEY+K+S + L+
Sbjct: 5 VVFLLLLSLVVLLLPLYASAAARVGGWSPISNVTDPQVVEIGEFAVSEYNKRSESGLKFE 64
Query: 306 KVVSGDSQVVAGTNYRLVIAAKGGSRSAAASNYEALVYERSWEHFRNLTSFKPVH 142
VVSG++QVV+GTNYRL +AA G + NY A+V+++ W FRNLTSF+P +
Sbjct: 65 TVVSGETQVVSGTNYRLKVAANDG--DGVSKNYLAIVWDKPWMKFRNLTSFEPAN 117
>pir||T07822 cystein proteinase inhibitor - cucumber gi|3237190|dbj|BAA28867.1|
cystein proteinase inhibitor [Cucumis sativus]
Length = 96
Score = 94.7 bits (234), Expect = 5e-19
Identities = 45/88 (51%), Positives = 61/88 (69%)
Frame = -3
Query: 417 VGGYTPIKDLNDPHVKEIAHFAVSEYDKQSGAKLRLVKVVSGDSQVVAGTNYRLVIAAKG 238
+GGY P KD NDPHVK+IA +AV+EY+K G L LV ++ +SQVVAG N+RLV+ K
Sbjct: 6 IGGYVPCKDPNDPHVKDIAEWAVAEYNKSQGHHLTLVSILKCESQVVAGVNWRLVLKCK- 64
Query: 237 GSRSAAASNYEALVYERSWEHFRNLTSF 154
+ NYE +V+E+ WE+FR L +F
Sbjct: 65 -DENNGEGNYETVVWEKIWENFRQLITF 91
>ref|NP_193383.1| cysteine proteinase inhibitor like protein; protein id:
At4g16500.1, supported by cDNA: 31946. [Arabidopsis
thaliana] gi|7438231|pir||H71431 hypothetical protein -
Arabidopsis thaliana gi|2245006|emb|CAB10426.1| cysteine
proteinase inhibitor like protein [Arabidopsis thaliana]
gi|7268400|emb|CAB78692.1| cysteine proteinase inhibitor
like protein [Arabidopsis thaliana]
gi|21592732|gb|AAM64681.1| cysteine proteinase inhibitor
like protein [Arabidopsis thaliana]
Length = 117
Score = 85.5 bits (210), Expect = 3e-16
Identities = 48/112 (42%), Positives = 68/112 (59%)
Frame = -3
Query: 486 IVSLLILSAVSGFSAALRQKGAFVGGYTPIKDLNDPHVKEIAHFAVSEYDKQSGAKLRLV 307
++ L ++S V G G +G PIK+++DP V +A +A+ E++K+S KL V
Sbjct: 11 LILLPLVSVVEGLGG-----GGGLGSRKPIKNVSDPDVVAVAKYAIEEHNKESKEKLVFV 65
Query: 306 KVVSGDSQVVAGTNYRLVIAAKGGSRSAAASNYEALVYERSWEHFRNLTSFK 151
KVV G +QVV+GT Y L IAAK G NYEA+V E+ W H ++L SFK
Sbjct: 66 KVVEGTTQVVSGTKYDLKIAAKDG--GGKIKNYEAVVVEKLWLHSKSLESFK 115
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 450,350,901
Number of Sequences: 1393205
Number of extensions: 10075148
Number of successful extensions: 33419
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 31801
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33274
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15942513235
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)