Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004051A_C01 KMC004051A_c01
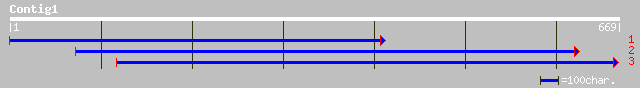
(669 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_177592.1| AtHVA22a; protein id: At1g74520.1, supported by... 96 4e-19
ref|NP_177128.1| AtHVA22c; protein id: At1g69700.1, supported by... 67 2e-10
ref|NP_201055.1| AtHVA22b-like protein; protein id: At5g62490.1,... 61 2e-08
gb|AAD31880.1| AtHVA22b [Arabidopsis thaliana] 61 2e-08
gb|EAA05741.1| agCP10297 [Anopheles gambiae str. PEST] 35 1.2
>ref|NP_177592.1| AtHVA22a; protein id: At1g74520.1, supported by cDNA: gi_4884931
[Arabidopsis thaliana] gi|25406352|pir||C96774 AtHVA22a,
65476-64429 [imported] - Arabidopsis thaliana
gi|4884932|gb|AAD31879.1| AtHVA22a [Arabidopsis
thaliana] gi|4884944|gb|AAD31885.1|AF141977_1 AtHVA22a
[Arabidopsis thaliana]
gi|12324799|gb|AAG52361.1|AC011765_13 AtHVA22a;
65476-64429 [Arabidopsis thaliana]
gi|26452664|dbj|BAC43415.1| putative AtHVA22a
[Arabidopsis thaliana] gi|28973175|gb|AAO63912.1|
putative AtHVA22a protein [Arabidopsis thaliana]
Length = 177
Score = 95.9 bits (237), Expect = 4e-19
Identities = 44/81 (54%), Positives = 56/81 (68%), Gaps = 6/81 (7%)
Frame = -3
Query: 667 SYVYEHYVRAFLLXPQTINIWYVPRKKDIFSKPDDIITAAEKYIQENGTQEFENLIHRAG 488
+YVYEH+VR + P++INIWYVP+K DIF KPDD++TAAEKYI ENG FE ++ RA
Sbjct: 95 AYVYEHFVRPVFVNPRSINIWYVPKKMDIFRKPDDVLTAAEKYIAENGPDAFEKILSRAD 154
Query: 487 KSRS------DGYHAMYDETY 443
KS+ + Y MY E Y
Sbjct: 155 KSKRYNKHEYESYETMYGEGY 175
>ref|NP_177128.1| AtHVA22c; protein id: At1g69700.1, supported by cDNA: 108964.,
supported by cDNA: gi_17529339, supported by cDNA:
gi_4884935 [Arabidopsis thaliana]
gi|25404831|pir||H96718 AtHVA22c, 50565-49239 [imported]
- Arabidopsis thaliana gi|4884936|gb|AAD31881.1|
AtHVA22c [Arabidopsis thaliana]
gi|4884946|gb|AAD31886.1|AF141978_1 AtHVA22c
[Arabidopsis thaliana]
gi|12325187|gb|AAG52538.1|AC013289_5 AtHVA22c;
50565-49239 [Arabidopsis thaliana]
gi|17529340|gb|AAL38897.1| putative AtHVA22c protein
[Arabidopsis thaliana] gi|21536712|gb|AAM61044.1|
AtHVA22c [Arabidopsis thaliana]
gi|23296865|gb|AAN13190.1| putative AtHVA22c protein
[Arabidopsis thaliana]
Length = 184
Score = 67.0 bits (162), Expect = 2e-10
Identities = 36/85 (42%), Positives = 52/85 (60%), Gaps = 11/85 (12%)
Frame = -3
Query: 664 YVYEHYVRAFLLXPQ--TINIWYVPRKK-DIFSK--PDDIITAAEKYIQENGTQEFENLI 500
++Y+H++R F PQ T IWYVP KK + F K DDI+TAAEKY++++GT+ FE +I
Sbjct: 98 HIYKHFIRPFYRDPQRATTKIWYVPHKKFNFFPKRDDDDILTAAEKYMEQHGTEAFERMI 157
Query: 499 ------HRAGKSRSDGYHAMYDETY 443
R SR H ++D+ Y
Sbjct: 158 VKKDSYERGRSSRGINNHMIFDDDY 182
>ref|NP_201055.1| AtHVA22b-like protein; protein id: At5g62490.1, supported by cDNA:
gi_4884933 [Arabidopsis thaliana]
gi|4884942|gb|AAD31884.1|AF141980_1 AtHVA22b
[Arabidopsis thaliana] gi|10178080|dbj|BAB11499.1|
AtHVA22b-like protein [Arabidopsis thaliana]
gi|26451511|dbj|BAC42853.1| putative AtHVA22b
[Arabidopsis thaliana] gi|28973349|gb|AAO63999.1|
putative AtHVA22b protein [Arabidopsis thaliana]
Length = 167
Score = 60.8 bits (146), Expect = 2e-08
Identities = 30/77 (38%), Positives = 46/77 (58%), Gaps = 2/77 (2%)
Frame = -3
Query: 667 SYVYEHYVRAFLLXPQTINIWYVPRKKDIFSKPDDIITAAEKY--IQENGTQEFENLIHR 494
+Y+YEHYVR+FLL P T+N+WYVP KKD DD+ A K+ + ++G + E ++
Sbjct: 95 AYLYEHYVRSFLLSPHTVNVWYVPAKKD-----DDLGATAGKFTPVNDSGAPQ-EKIVSS 148
Query: 493 AGKSRSDGYHAMYDETY 443
S H+ +D+ Y
Sbjct: 149 VDTSAKYVGHSAFDDAY 165
>gb|AAD31880.1| AtHVA22b [Arabidopsis thaliana]
Length = 167
Score = 60.8 bits (146), Expect = 2e-08
Identities = 30/77 (38%), Positives = 46/77 (58%), Gaps = 2/77 (2%)
Frame = -3
Query: 667 SYVYEHYVRAFLLXPQTINIWYVPRKKDIFSKPDDIITAAEKY--IQENGTQEFENLIHR 494
+Y+YEHYVR+FLL P T+N+WYVP KKD DD+ A K+ + ++G + E ++
Sbjct: 95 AYLYEHYVRSFLLSPHTVNVWYVPAKKD-----DDLGATAGKFTPVNDSGAPQ-EKIVSS 148
Query: 493 AGKSRSDGYHAMYDETY 443
S H+ +D+ Y
Sbjct: 149 VDTSAKYVGHSAFDDAY 165
>gb|EAA05741.1| agCP10297 [Anopheles gambiae str. PEST]
Length = 348
Score = 34.7 bits (78), Expect = 1.2
Identities = 29/102 (28%), Positives = 48/102 (46%), Gaps = 9/102 (8%)
Frame = +2
Query: 383 ILAYTCIPLSL*GRSYKNHSIGLIIHCMITIT-P*FTSPMDQILKFLCSILLYVFLGSSY 559
+L Y L L G SY+ + +++ IT+ P FT+ + F+CS+ VF+G+ +
Sbjct: 15 LLLYIQERLGLWGESYRARFLFVVVAFSITVCIPKFTTTYTNLETFICSMAELVFIGNVF 74
Query: 560 N---VIWLTEDVLFPWNIPDVNGL-----RXEQESSHIMLVN 661
++W TE F I VN L R + H++ N
Sbjct: 75 GGAMLLW-TEYDAFRQFIEQVNSLTKHFYREDPLKEHVLQFN 115
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 549,162,114
Number of Sequences: 1393205
Number of extensions: 11653945
Number of successful extensions: 24244
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 20015
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24178
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29138478756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)