Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004048A_C01 KMC004048A_c01
(596 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
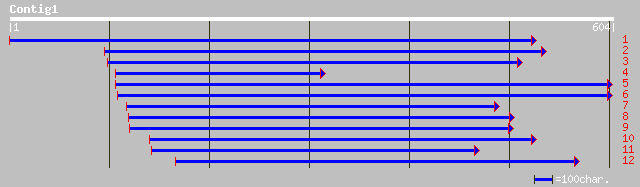
dbj|BAA89236.1| TMV response-related gene product [Nicotiana tab... 95 8e-19
dbj|BAC42824.1| unknown protein [Arabidopsis thaliana] 87 2e-16
ref|NP_186785.1| hypothetical protein; protein id: At3g01360.1 [... 87 2e-16
pir||S14183 DNA-directed RNA polymerase (EC 2.7.7.6) largest cha... 33 3.6
gb|AAK13048.1| paraquat-inducible protein [Burkholderia pseudoma... 32 4.7
>dbj|BAA89236.1| TMV response-related gene product [Nicotiana tabacum]
Length = 222
Score = 94.7 bits (234), Expect = 8e-19
Identities = 54/103 (52%), Positives = 68/103 (65%), Gaps = 8/103 (7%)
Frame = -1
Query: 596 EYHRARAIATLQFNCHLALMVMVMVASYSVFCGRNGGPVHPEASRYTPLGAEMQSLE--N 423
EYHR RAIATLQFNCHLAL+V V+ YS+ C +NG + PE RY P+GAEMQ LE +
Sbjct: 121 EYHRGRAIATLQFNCHLALLVTVIAFVYSIVCKKNG--IGPEHMRYRPIGAEMQHLEMDS 178
Query: 422 SATFTLDSDDEDIKDG-----NVGSQKGVVVV-EHGMNGNATN 312
FTL+SD++D ++G NV QK +V E G NG T+
Sbjct: 179 QGHFTLESDEDDDENGIKEERNVEMQKAIVTAPESGTNGYHTH 221
>dbj|BAC42824.1| unknown protein [Arabidopsis thaliana]
Length = 162
Score = 87.0 bits (214), Expect = 2e-16
Identities = 49/95 (51%), Positives = 65/95 (67%)
Frame = -1
Query: 596 EYHRARAIATLQFNCHLALMVMVMVASYSVFCGRNGGPVHPEASRYTPLGAEMQSLENSA 417
+YHRA+AIATLQFNCHLALMV+V +SV +N G + + S+Y PLGAE LEN +
Sbjct: 76 DYHRAKAIATLQFNCHLALMVVVATGLFSVIANKN-GYLRQDHSKYRPLGAE---LENLS 131
Query: 416 TFTLDSDDEDIKDGNVGSQKGVVVVEHGMNGNATN 312
TFTLDSD+ED ++ V E G+NGN+++
Sbjct: 132 TFTLDSDEED-----EVREESNVAKEVGLNGNSSH 161
>ref|NP_186785.1| hypothetical protein; protein id: At3g01360.1 [Arabidopsis
thaliana] gi|6094549|gb|AAF03491.1|AC010676_1
hypothetical protein [Arabidopsis thaliana]
Length = 319
Score = 87.0 bits (214), Expect = 2e-16
Identities = 49/95 (51%), Positives = 65/95 (67%)
Frame = -1
Query: 596 EYHRARAIATLQFNCHLALMVMVMVASYSVFCGRNGGPVHPEASRYTPLGAEMQSLENSA 417
+YHRA+AIATLQFNCHLALMV+V +SV +N G + + S+Y PLGAE LEN +
Sbjct: 233 DYHRAKAIATLQFNCHLALMVVVATGLFSVIANKN-GYLRQDHSKYRPLGAE---LENLS 288
Query: 416 TFTLDSDDEDIKDGNVGSQKGVVVVEHGMNGNATN 312
TFTLDSD+ED ++ V E G+NGN+++
Sbjct: 289 TFTLDSDEED-----EVREESNVAKEVGLNGNSSH 318
>pir||S14183 DNA-directed RNA polymerase (EC 2.7.7.6) largest chain (isoform C) -
soybean (fragment) gi|18736|emb|CAA36736.1| DNA-directed
RNA polymerase [Glycine max]
Length = 977
Score = 32.7 bits (73), Expect = 3.6
Identities = 20/67 (29%), Positives = 29/67 (42%)
Frame = +1
Query: 325 PFIPCSTTTTPFWLPTLPSLMSSSSESNVKVAEFSKDCISAPSGVYLEASG*TGPPFLPQ 504
P P + T+P + PT PS +S N + A++S +PS L T P + P
Sbjct: 852 PTSPAYSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSP---TSPNYSPT 908
Query: 505 KTEYEAT 525
Y T
Sbjct: 909 SPSYSPT 915
>gb|AAK13048.1| paraquat-inducible protein [Burkholderia pseudomallei]
Length = 553
Score = 32.3 bits (72), Expect = 4.7
Identities = 31/116 (26%), Positives = 48/116 (40%), Gaps = 26/116 (22%)
Frame = +1
Query: 181 NFIHKMKNYNMIFLYNMIPNKLG----EGGRDKLQKGKRDCIGQ---------------- 300
+F K KN+ M N+ P++LG E +DK +R+ + +
Sbjct: 346 DFDPKTKNFTMPVTMNVYPDRLGRKFREVSQDKGYAARREVLSRLVQHGLRGQLRTGNLL 405
Query: 301 TSQ*LVALPFIP------CSTTTTPFWLPTLPSLMSSSSESNVKVAEFSKDCISAP 450
TSQ VAL F P P LPT+P+ + E ++VA+ +K P
Sbjct: 406 TSQLYVALDFFPKAPPVKIDLAREPVELPTVPNTL---DELQLQVADIAKKLDKVP 458
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 505,795,363
Number of Sequences: 1393205
Number of extensions: 10628414
Number of successful extensions: 31641
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 30102
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31597
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)