Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004046A_C01 KMC004046A_c01
(922 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
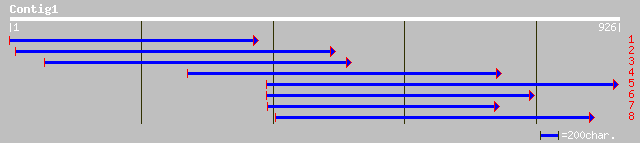
ref|NP_177395.1| unknown protein; protein id: At1g72510.1 [Arabi... 37 0.42
ref|NP_178825.1| unknown protein; protein id: At2g09970.1 [Arabi... 36 0.72
ref|NP_197567.1| putative protein; protein id: At5g20670.1, supp... 36 0.94
gb|AAM63436.1| unknown [Arabidopsis thaliana] 35 1.2
emb|CAD61084.1| SI:dZ46M7.2 (novel kinase) [Danio rerio] 35 2.1
>ref|NP_177395.1| unknown protein; protein id: At1g72510.1 [Arabidopsis thaliana]
gi|25406187|pir||D96749 unknown protein T10D10.2
[imported] - Arabidopsis thaliana
gi|12325270|gb|AAG52577.1|AC016529_8 unknown protein;
9323-8826 [Arabidopsis thaliana]
Length = 165
Score = 37.0 bits (84), Expect = 0.42
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 19/73 (26%)
Frame = -1
Query: 916 RHIGFCQEFRYSAPNEED---LVHAVKQILFRSLDSPRKER--------------FSC-- 794
RH+ C +F+ S+P L+ A++QIL +SLDSPR R C
Sbjct: 92 RHMNMCNKFKSSSPPPNPTGHLISAMRQILRKSLDSPRMLRSMPNSPSKDDQDHHHDCVS 151
Query: 793 RPLGRSQSCFSTM 755
L RS SCF+++
Sbjct: 152 NVLSRSDSCFASL 164
>ref|NP_178825.1| unknown protein; protein id: At2g09970.1 [Arabidopsis thaliana]
gi|25411336|pir||D84489 hypothetical protein At2g09970
[imported] - Arabidopsis thaliana
gi|4558678|gb|AAD22695.1| unknown protein [Arabidopsis
thaliana]
Length = 166
Score = 36.2 bits (82), Expect = 0.72
Identities = 25/73 (34%), Positives = 36/73 (49%), Gaps = 19/73 (26%)
Frame = -1
Query: 916 RHIGFCQEFRYSAPNEE---DLVHAVKQILFRSLDSPRKER--------------FSC-- 794
RH+ C +F+ S+P L+ A++QIL +SLDSPR R C
Sbjct: 93 RHMNKCNKFKSSSPPSNLTRHLISAMRQILRKSLDSPRMLRSMPNSPSKDDQDNHHDCVS 152
Query: 793 RPLGRSQSCFSTM 755
L RS SCF+++
Sbjct: 153 NVLSRSDSCFASL 165
>ref|NP_197567.1| putative protein; protein id: At5g20670.1, supported by cDNA:
250877. [Arabidopsis thaliana]
Length = 153
Score = 35.8 bits (81), Expect = 0.94
Identities = 28/73 (38%), Positives = 35/73 (47%), Gaps = 17/73 (23%)
Frame = -1
Query: 922 LDRHIGFCQEFR-YSAPNEEDLVHAVKQILFRSLD-SPRKERF---------------SC 794
L RH FC FR +S EED + + +IL RSLD SPR+ S
Sbjct: 79 LRRHTTFCHRFRSWSPDEEEDPISVIGRILRRSLDGSPRRTNTRTSSSGALPGIDGVESR 138
Query: 793 RPLGRSQSCFSTM 755
R L RS SCF ++
Sbjct: 139 RSLLRSGSCFPSL 151
>gb|AAM63436.1| unknown [Arabidopsis thaliana]
Length = 153
Score = 35.4 bits (80), Expect = 1.2
Identities = 28/73 (38%), Positives = 35/73 (47%), Gaps = 17/73 (23%)
Frame = -1
Query: 922 LDRHIGFCQEFR-YSAPNEEDLVHAVKQILFRSLD-SPRKERF---------------SC 794
L RH FC FR +S EED + + +IL RSLD SPR+ S
Sbjct: 79 LRRHTTFCHRFRSWSPDEEEDPISVIGRILRRSLDGSPRRTTTRTSSSGALPGIDGVESR 138
Query: 793 RPLGRSQSCFSTM 755
R L RS SCF ++
Sbjct: 139 RSLLRSGSCFPSL 151
>emb|CAD61084.1| SI:dZ46M7.2 (novel kinase) [Danio rerio]
Length = 553
Score = 34.7 bits (78), Expect = 2.1
Identities = 21/57 (36%), Positives = 24/57 (41%)
Frame = -1
Query: 892 FRYSAPNEEDLVHAVKQILFRSLDSPRKERFSCRPLGRSQSCFSTMQVTTPPPESPS 722
F YS PN ED + Q + LD CRP RS C ST Q P P+
Sbjct: 29 FNYSLPNPEDAQEYLAQSAWPELDGVSS---FCRPAARSLLCHSTFQDCNPSGLGPA 82
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 709,793,829
Number of Sequences: 1393205
Number of extensions: 14768850
Number of successful extensions: 33954
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 31071
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33691
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 50750480856
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)