Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
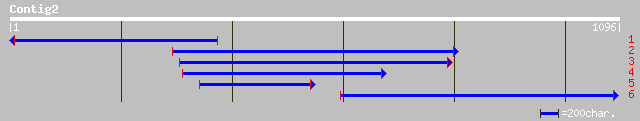
Query= KMC004033A_C02 KMC004033A_c02
(1093 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_196645.1| endomembrane protein 70, putative; protein id: ... 86 1e-15
ref|NP_568465.1| endomembrane protein 70, putative; protein id: ... 83 9e-15
emb|CAC47950.1| PHG1A protein [Dictyostelium discoideum] 64 8e-14
ref|NP_179994.1| endomembrane protein 70, putative; protein id: ... 79 1e-13
gb|AAM13887.1| putative multispanning membrane protein [Arabidop... 79 1e-13
>ref|NP_196645.1| endomembrane protein 70, putative; protein id: At5g10840.1
[Arabidopsis thaliana] gi|11281523|pir||T50793
hypothetical protein T30N20_110 - Arabidopsis thaliana
gi|8979718|emb|CAB96839.1| putative protein [Arabidopsis
thaliana]
Length = 639
Score = 85.9 bits (211), Expect = 1e-15
Identities = 48/114 (42%), Positives = 65/114 (56%), Gaps = 21/114 (18%)
Frame = -1
Query: 766 FSLVFLWFGISVPLVFVGGHIDFKKKVI*DLVETSKIPWQIPEQHWYMNSSYSLLL---- 599
F+L+FLWFGISVPLVFVGG+I FKK D V+T+KIP QIPEQ WYMN +S+L+
Sbjct: 447 FALIFLWFGISVPLVFVGGYIGFKKPAADDPVKTNKIPRQIPEQAWYMNPVFSILIGGIL 506
Query: 598 ---------------VLRSQYFFATSSCVVFFFSTFVPC--ISALLCIG*LCSQ 488
+ +Q+++ + F V C I+ +LC LCS+
Sbjct: 507 PFGAVFIELFFILTSIWLNQFYYIFGFLFLVFVILIVTCAEITVVLCYFQLCSE 560
Score = 44.3 bits (103), Expect = 0.003
Identities = 25/64 (39%), Positives = 36/64 (56%), Gaps = 3/64 (4%)
Frame = -3
Query: 617 IILIVTCAAITILLCYFQLCSVFF*YFCSLYF---SSVVYWIVMFSSLVVLILTTSKLAG 447
+ILIVTCA IT++LCYFQLCS + ++ Y SS +Y + + L +KL
Sbjct: 539 VILIVTCAEITVVLCYFQLCSEDYLWWWRSYLTSGSSALYLFLYATFYFFTKLQITKLVS 598
Query: 446 ILEY 435
+ Y
Sbjct: 599 AMLY 602
>ref|NP_568465.1| endomembrane protein 70, putative; protein id: At5g25100.1,
supported by cDNA: gi_13430445 [Arabidopsis thaliana]
gi|13430446|gb|AAK25845.1|AF360135_1 putative
multispanning membrane protein [Arabidopsis thaliana]
Length = 644
Score = 82.8 bits (203), Expect = 9e-15
Identities = 46/114 (40%), Positives = 65/114 (56%), Gaps = 21/114 (18%)
Frame = -1
Query: 766 FSLVFLWFGISVPLVFVGGHIDFKKKVI*DLVETSKIPWQIPEQHWYMNSSYSLLL---- 599
F+L+FLWFGISVPLVFVG ++ FKK + D V+T+KIP QIPEQ WYMN +S+L+
Sbjct: 452 FALIFLWFGISVPLVFVGAYLGFKKPPLDDPVKTNKIPRQIPEQAWYMNPIFSILIGGIL 511
Query: 598 ---------------VLRSQYFFATSSCVVFFFSTFVPC--ISALLCIG*LCSQ 488
+ +Q+++ + F V C I+ +LC LCS+
Sbjct: 512 PFGAVFIELFFILTSIWLNQFYYIFGFLFLVFVILMVTCAEITIVLCYFQLCSE 565
Score = 44.7 bits (104), Expect = 0.003
Identities = 26/64 (40%), Positives = 36/64 (55%), Gaps = 3/64 (4%)
Frame = -3
Query: 617 IILIVTCAAITILLCYFQLCSVFF*YFCSLYF---SSVVYWIVMFSSLVVLILTTSKLAG 447
+IL+VTCA ITI+LCYFQLCS + ++ Y SS VY + + L +KL
Sbjct: 544 VILMVTCAEITIVLCYFQLCSEDYLWWWRSYLTSGSSAVYLFLYAAFYFFTKLQITKLVS 603
Query: 446 ILEY 435
+ Y
Sbjct: 604 AMLY 607
>emb|CAC47950.1| PHG1A protein [Dictyostelium discoideum]
Length = 641
Score = 63.9 bits (154), Expect(2) = 8e-14
Identities = 29/55 (52%), Positives = 40/55 (72%)
Frame = -1
Query: 763 SLVFLWFGISVPLVFVGGHIDFKKKVI*DLVETSKIPWQIPEQHWYMNSSYSLLL 599
S++ +WFGISVPLVF+G + KK V D V T++IP Q+P+Q WYMN S+L+
Sbjct: 451 SIIAMWFGISVPLVFLGSYFASKKPVPEDPVRTNQIPRQVPDQIWYMNPYLSILM 505
Score = 35.8 bits (81), Expect(2) = 8e-14
Identities = 15/22 (68%), Positives = 20/22 (90%)
Frame = -3
Query: 620 LIILIVTCAAITILLCYFQLCS 555
L+ILIVT A I+I++CYFQLC+
Sbjct: 541 LMILIVTSAEISIVMCYFQLCA 562
>ref|NP_179994.1| endomembrane protein 70, putative; protein id: At2g24170.1
[Arabidopsis thaliana] gi|25346720|pir||D84633 probable
multispanning membrane protein [imported] - Arabidopsis
thaliana gi|4115377|gb|AAD03378.1| putative
multispanning membrane protein [Arabidopsis thaliana]
Length = 659
Score = 79.3 bits (194), Expect = 1e-13
Identities = 46/114 (40%), Positives = 64/114 (55%), Gaps = 21/114 (18%)
Frame = -1
Query: 766 FSLVFLWFGISVPLVFVGGHIDFKKKVI*DLVETSKIPWQIPEQHWYMNSSYSLLL---- 599
F+LV LWFGISVPLVF+GG+I F+K D V+T+KIP QIP Q WYMN +S+L+
Sbjct: 467 FALVVLWFGISVPLVFIGGYIGFRKPAPEDPVKTNKIPRQIPTQAWYMNPIFSILIGGIL 526
Query: 598 -----------VLRS----QYFFATSSCVVFFFSTFVPC--ISALLCIG*LCSQ 488
+L S Q+++ + F + C I+ +LC LCS+
Sbjct: 527 PFGAVFIELFFILTSIWLHQFYYIFGFLFIVFIILIITCAEITVVLCYFQLCSE 580
Score = 44.3 bits (103), Expect = 0.003
Identities = 22/59 (37%), Positives = 35/59 (59%)
Frame = -3
Query: 617 IILIVTCAAITILLCYFQLCSVFF*YFCSLYFSSVVYWIVMFSSLVVLILTTSKLAGIL 441
IILI+TCA IT++LCYFQLCS + ++ Y +S + +F V T ++ ++
Sbjct: 559 IILIITCAEITVVLCYFQLCSEDYQWWWRSYLTSGSSAVYLFLYAVFYFYTKLEITKLV 617
>gb|AAM13887.1| putative multispanning membrane protein [Arabidopsis thaliana]
Length = 637
Score = 79.3 bits (194), Expect = 1e-13
Identities = 46/114 (40%), Positives = 64/114 (55%), Gaps = 21/114 (18%)
Frame = -1
Query: 766 FSLVFLWFGISVPLVFVGGHIDFKKKVI*DLVETSKIPWQIPEQHWYMNSSYSLLL---- 599
F+LV LWFGISVPLVF+GG+I F+K D V+T+KIP QIP Q WYMN +S+L+
Sbjct: 445 FALVVLWFGISVPLVFIGGYIGFRKPAPEDPVKTNKIPRQIPTQAWYMNPIFSILIGGIL 504
Query: 598 -----------VLRS----QYFFATSSCVVFFFSTFVPC--ISALLCIG*LCSQ 488
+L S Q+++ + F + C I+ +LC LCS+
Sbjct: 505 PFGAVFIELFFILTSIWLHQFYYIFGFLFIVFIILIITCAEITVVLCYFQLCSE 558
Score = 44.3 bits (103), Expect = 0.003
Identities = 22/59 (37%), Positives = 35/59 (59%)
Frame = -3
Query: 617 IILIVTCAAITILLCYFQLCSVFF*YFCSLYFSSVVYWIVMFSSLVVLILTTSKLAGIL 441
IILI+TCA IT++LCYFQLCS + ++ Y +S + +F V T ++ ++
Sbjct: 537 IILIITCAEITVVLCYFQLCSEDYQWWWRSYLTSGSSAVYLFLYAVFYFYTKLEITKLV 595
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 919,876,393
Number of Sequences: 1393205
Number of extensions: 19993590
Number of successful extensions: 55971
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 53032
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 55941
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 65340192036
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)