Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004032A_C01 KMC004032A_c01
(550 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
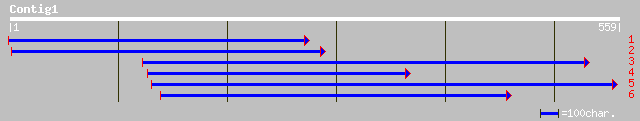
ref|NP_176711.1| hypothetical protein; protein id: At1g65320.1 [... 122 2e-27
pir||T02355 hypothetical protein T8F5.10 - Arabidopsis thaliana ... 122 2e-27
dbj|BAB86114.1| P0678F11.21 [Oryza sativa (japonica cultivar-gro... 101 7e-21
gb|AAM93687.1| unknown protein [Oryza sativa (japonica cultivar-... 51 1e-05
gb|AAO22709.1| unknown protein [Arabidopsis thaliana] 41 0.011
>ref|NP_176711.1| hypothetical protein; protein id: At1g65320.1 [Arabidopsis
thaliana] gi|26449327|dbj|BAC41791.1| unknown protein
[Arabidopsis thaliana]
Length = 425
Score = 122 bits (307), Expect = 2e-27
Identities = 59/74 (79%), Positives = 65/74 (87%)
Frame = -3
Query: 509 RSMYRGRSAPLTCKITSSLAAVMAQMLSHRATHVWVTEDESDDVIVGVVGYADIFAAVTK 330
RSMYRGRSAPLTCK +SSLAAVMAQMLSHRATHVWVTE +SDDV+VGVVGY +I AVTK
Sbjct: 345 RSMYRGRSAPLTCKTSSSLAAVMAQMLSHRATHVWVTEADSDDVLVGVVGYGEILTAVTK 404
Query: 329 PPTTFTPANRSTEG 288
P+ F P+NRS EG
Sbjct: 405 QPSAFVPSNRSYEG 418
>pir||T02355 hypothetical protein T8F5.10 - Arabidopsis thaliana
gi|3335341|gb|AAC27143.1|AAC27143 T8F5.10 [Arabidopsis
thaliana]
Length = 482
Score = 122 bits (307), Expect = 2e-27
Identities = 59/74 (79%), Positives = 65/74 (87%)
Frame = -3
Query: 509 RSMYRGRSAPLTCKITSSLAAVMAQMLSHRATHVWVTEDESDDVIVGVVGYADIFAAVTK 330
RSMYRGRSAPLTCK +SSLAAVMAQMLSHRATHVWVTE +SDDV+VGVVGY +I AVTK
Sbjct: 402 RSMYRGRSAPLTCKTSSSLAAVMAQMLSHRATHVWVTEADSDDVLVGVVGYGEILTAVTK 461
Query: 329 PPTTFTPANRSTEG 288
P+ F P+NRS EG
Sbjct: 462 QPSAFVPSNRSYEG 475
>dbj|BAB86114.1| P0678F11.21 [Oryza sativa (japonica cultivar-group)]
Length = 404
Score = 101 bits (251), Expect = 7e-21
Identities = 57/77 (74%), Positives = 60/77 (77%), Gaps = 4/77 (5%)
Frame = -3
Query: 548 GFFSNSASP-GFGS-RSMYRGRSAPLTCKITSSLAAVMAQMLSHRATHVWVT--EDESDD 381
GF ++ A FG RSMYRGRSAPL CK TSSLAAVMAQMLSHRATHVWVT E E D
Sbjct: 321 GFLNSQAHQMAFGRMRSMYRGRSAPLMCKSTSSLAAVMAQMLSHRATHVWVTDAESEEDG 380
Query: 380 VIVGVVGYADIFAAVTK 330
V+VGVVGY DIF AVTK
Sbjct: 381 VLVGVVGYTDIFNAVTK 397
>gb|AAM93687.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 385
Score = 50.8 bits (120), Expect = 1e-05
Identities = 21/50 (42%), Positives = 33/50 (66%)
Frame = -3
Query: 497 RGRSAPLTCKITSSLAAVMAQMLSHRATHVWVTEDESDDVIVGVVGYADI 348
R P+ C SSL AVM Q L+HRA+++WV +++ D + G+V +AD+
Sbjct: 327 RSTEEPVVCSPASSLVAVMMQALAHRASYLWVLDEDDDCRLAGIVTFADV 376
>gb|AAO22709.1| unknown protein [Arabidopsis thaliana]
Length = 391
Score = 40.8 bits (94), Expect = 0.011
Identities = 21/54 (38%), Positives = 28/54 (50%)
Frame = -3
Query: 491 RSAPLTCKITSSLAAVMAQMLSHRATHVWVTEDESDDVIVGVVGYADIFAAVTK 330
+S + C SSL AVM Q ++HR + WV E D VG+V + DI K
Sbjct: 334 KSEAIVCNPKSSLMAVMIQAVAHRVNYAWVV--EKDGCFVGMVTFVDILKVFRK 385
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,107,064
Number of Sequences: 1393205
Number of extensions: 10420756
Number of successful extensions: 27499
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 26086
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27417
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)