Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004029A_C01 KMC004029A_c01
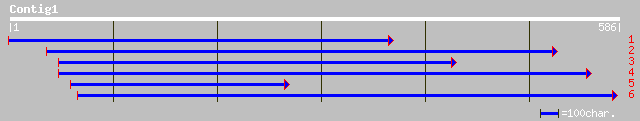
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_171661.1| protein serine/threonine kinase, putative; prot... 60 2e-08
gb|AAK56254.1|AF367265_1 At1g01540/F22L4_6 [Arabidopsis thaliana... 60 2e-08
pir||A86146 hypothetical protein F22L4.8 - Arabidopsis thaliana ... 60 2e-08
ref|NP_192172.1| putative serine/threonine protein kinase; prote... 60 2e-08
dbj|BAB90415.1| similar to protein kinase [Oryza sativa (japonic... 59 6e-08
>ref|NP_171661.1| protein serine/threonine kinase, putative; protein id: At1g01540.1,
supported by cDNA: gi_12083265, supported by cDNA:
gi_14194118, supported by cDNA: gi_20334727 [Arabidopsis
thaliana] gi|2505874|emb|CAA73303.1| putative kinase
[Arabidopsis thaliana]
gi|12083266|gb|AAG48792.1|AF332429_1 putative protein
serine/threonine kinase [Arabidopsis thaliana]
Length = 472
Score = 60.5 bits (145), Expect = 2e-08
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = -3
Query: 584 RCTDPNAQKRPKMGHVIHMLEAEDSPYKEDRRARRD 477
RC DP+A KRPKMGH+IHMLEAED Y+++RR RD
Sbjct: 406 RCVDPDANKRPKMGHIIHMLEAEDLLYRDERRTTRD 441
>gb|AAK56254.1|AF367265_1 At1g01540/F22L4_6 [Arabidopsis thaliana] gi|20334728|gb|AAM16225.1|
At1g01540/F22L4_6 [Arabidopsis thaliana]
Length = 479
Score = 60.5 bits (145), Expect = 2e-08
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = -3
Query: 584 RCTDPNAQKRPKMGHVIHMLEAEDSPYKEDRRARRD 477
RC DP+A KRPKMGH+IHMLEAED Y+++RR RD
Sbjct: 406 RCVDPDANKRPKMGHIIHMLEAEDLLYRDERRTTRD 441
>pir||A86146 hypothetical protein F22L4.8 - Arabidopsis thaliana
gi|8920590|gb|AAF81312.1|AC061957_8 Contains a strong
similarity to an unknown protein from Arabidopsis
thaliana gi|2505874 and contains an eukaryotic protein
kinase PF|00069 domain. ESTs gb|Z26473, gb|AI996016,
gb|Z17558, gb|N97089, gb|BE039500, gb|AA712856,
gb|Z26772 come from this gene
Length = 497
Score = 60.5 bits (145), Expect = 2e-08
Identities = 25/36 (69%), Positives = 30/36 (82%)
Frame = -3
Query: 584 RCTDPNAQKRPKMGHVIHMLEAEDSPYKEDRRARRD 477
RC DP+A KRPKMGH+IHMLEAED Y+++RR RD
Sbjct: 431 RCVDPDANKRPKMGHIIHMLEAEDLLYRDERRTTRD 466
>ref|NP_192172.1| putative serine/threonine protein kinase; protein id: At4g02630.1
[Arabidopsis thaliana] gi|7488204|pir||T01086 probable
serine/threonine-specific protein kinase (EC 2.7.1.-)
T10P11.10 - Arabidopsis thaliana
gi|2262143|gb|AAC78256.1|AAC78256 putative
serine/threonine protein kinase [Arabidopsis thaliana]
gi|7269023|emb|CAB80756.1| putative serine/threonine
protein kinase [Arabidopsis thaliana]
gi|28393613|gb|AAO42226.1| putative serine/threonine
protein kinase [Arabidopsis thaliana]
gi|28973357|gb|AAO64003.1| putative serine/threonine
protein kinase [Arabidopsis thaliana]
Length = 492
Score = 60.1 bits (144), Expect = 2e-08
Identities = 26/32 (81%), Positives = 28/32 (87%)
Frame = -3
Query: 584 RCTDPNAQKRPKMGHVIHMLEAEDSPYKEDRR 489
RC DPNAQKRPKMGH+IHMLEAED K+DRR
Sbjct: 415 RCVDPNAQKRPKMGHIIHMLEAEDLVSKDDRR 446
>dbj|BAB90415.1| similar to protein kinase [Oryza sativa (japonica cultivar-group)]
Length = 411
Score = 58.5 bits (140), Expect = 6e-08
Identities = 25/43 (58%), Positives = 33/43 (76%)
Frame = -3
Query: 584 RCTDPNAQKRPKMGHVIHMLEAEDSPYKEDRRARRDTVQSPNI 456
RC DP+++KRPKMG V+ MLE+E+ PY+EDRR RR S +I
Sbjct: 340 RCVDPDSEKRPKMGQVVRMLESEEVPYREDRRNRRSRTGSMDI 382
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,943,456
Number of Sequences: 1393205
Number of extensions: 10687622
Number of successful extensions: 23708
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 23009
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23691
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)