Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004008A_C01 KMC004008A_c01
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
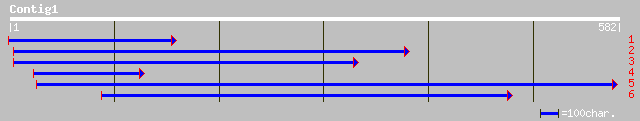
gb|AAB82764.1| b-keto acyl reductase [Allium porrum] 181 4e-45
pir||T06214 probable b-keto acyl reductase (EC 1.1.-.-) - barley... 172 3e-42
pir||T02061 probable beta-keto acyl reductase - maize gi|2586129... 170 1e-41
gb|AAO43449.1| putative 3-ketoacyl-CoA reductase 2 [Brassica napus] 154 1e-36
gb|AAO43448.1| putative 3-ketoacyl-CoA reductase 1 [Brassica napus] 153 2e-36
>gb|AAB82764.1| b-keto acyl reductase [Allium porrum]
Length = 229
Score = 181 bits (460), Expect = 4e-45
Identities = 79/111 (71%), Positives = 96/111 (86%)
Frame = -2
Query: 576 ATKAYIDQFSRCLYVEYKKSGIDVQCQVPLYVATKMASIKKSSFFVPSTDGYARAGVRWI 397
ATKAY+DQFSRCL+VEYK GIDVQCQVPLYVATKMASI++SSF VPS+D YA+A +RWI
Sbjct: 118 ATKAYVDQFSRCLFVEYKSKGIDVQCQVPLYVATKMASIRRSSFMVPSSDTYAKAALRWI 177
Query: 396 GHDPRCTPYWPHTLLWALAASLPEFVIDAWRLRFCLAIRKRGQLKESRKKE 244
G++PRCTPYWPH+ +W L + LPE ID WRL FCL IR +GQLK+++KK+
Sbjct: 178 GYEPRCTPYWPHSAIWCLLSLLPESAIDNWRLGFCLGIRTKGQLKDAKKKD 228
>pir||T06214 probable b-keto acyl reductase (EC 1.1.-.-) - barley
gi|2586127|gb|AAB82766.1| b-keto acyl reductase [Hordeum
vulgare]
Length = 325
Score = 172 bits (436), Expect = 3e-42
Identities = 77/110 (70%), Positives = 94/110 (85%)
Frame = -2
Query: 576 ATKAYIDQFSRCLYVEYKKSGIDVQCQVPLYVATKMASIKKSSFFVPSTDGYARAGVRWI 397
ATKAY+DQFSRCLYVEYK GIDVQCQVPLYVATKMASI++SSF VPS D YARA +R I
Sbjct: 214 ATKAYVDQFSRCLYVEYKGKGIDVQCQVPLYVATKMASIRRSSFLVPSADTYARAAIRHI 273
Query: 396 GHDPRCTPYWPHTLLWALAASLPEFVIDAWRLRFCLAIRKRGQLKESRKK 247
G++PRCTPYWPH++LW L + LPE ++D+ RL C+ IRK+GQ K+++KK
Sbjct: 274 GYEPRCTPYWPHSVLWFLISLLPESLVDSTRLSMCIKIRKKGQAKDAKKK 323
>pir||T02061 probable beta-keto acyl reductase - maize gi|2586129|gb|AAB82767.1|
b-keto acyl reductase [Zea mays]
gi|15824366|gb|AAL09309.1|AF302098_1 beta-ketoacyl
reductase [Zea mays]
Length = 326
Score = 170 bits (431), Expect = 1e-41
Identities = 78/110 (70%), Positives = 93/110 (83%)
Frame = -2
Query: 576 ATKAYIDQFSRCLYVEYKKSGIDVQCQVPLYVATKMASIKKSSFFVPSTDGYARAGVRWI 397
ATKAY+DQFSRCLYVEYK GIDVQCQVPLYVATKMASI+KSSF VPS D YARA VR I
Sbjct: 215 ATKAYVDQFSRCLYVEYKSKGIDVQCQVPLYVATKMASIRKSSFMVPSADTYARAAVRHI 274
Query: 396 GHDPRCTPYWPHTLLWALAASLPEFVIDAWRLRFCLAIRKRGQLKESRKK 247
G++PRCTPYWPH+++W L + LPE +ID+ RL C+ IRK+G K+++KK
Sbjct: 275 GYEPRCTPYWPHSVVWFLISILPESLIDSVRLGMCIKIRKKGLAKDAKKK 324
>gb|AAO43449.1| putative 3-ketoacyl-CoA reductase 2 [Brassica napus]
Length = 319
Score = 154 bits (388), Expect = 1e-36
Identities = 66/109 (60%), Positives = 90/109 (82%)
Frame = -2
Query: 570 KAYIDQFSRCLYVEYKKSGIDVQCQVPLYVATKMASIKKSSFFVPSTDGYARAGVRWIGH 391
K Y+DQFSRCL+VEYKKSGIDVQCQVPLYVATKM I+++SF V S +GYA+A +R++G+
Sbjct: 210 KTYVDQFSRCLHVEYKKSGIDVQCQVPLYVATKMTKIRRASFLVASPEGYAKAALRFVGY 269
Query: 390 DPRCTPYWPHTLLWALAASLPEFVIDAWRLRFCLAIRKRGQLKESRKKE 244
+PRCTPYWPH L+ + ++LPE V +++ ++ CL IRK+G LK+S K+
Sbjct: 270 EPRCTPYWPHALMGYVVSALPESVFESFNIKRCLQIRKKGMLKDSSSKK 318
>gb|AAO43448.1| putative 3-ketoacyl-CoA reductase 1 [Brassica napus]
Length = 319
Score = 153 bits (386), Expect = 2e-36
Identities = 66/109 (60%), Positives = 90/109 (82%)
Frame = -2
Query: 570 KAYIDQFSRCLYVEYKKSGIDVQCQVPLYVATKMASIKKSSFFVPSTDGYARAGVRWIGH 391
K Y+DQFSRCL+VEYKKSGIDVQCQVPLYVATKM I+++SF V S +GYA+A +R++G+
Sbjct: 210 KTYVDQFSRCLHVEYKKSGIDVQCQVPLYVATKMTKIRRASFLVASPEGYAKAALRFVGY 269
Query: 390 DPRCTPYWPHTLLWALAASLPEFVIDAWRLRFCLAIRKRGQLKESRKKE 244
+PRCTPYWPH L+ + ++LPE V +++ ++ CL IRK+G LK+S K+
Sbjct: 270 EPRCTPYWPHALMGYVVSALPESVFESFNIKRCLQIRKKGMLKDSISKK 318
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,824,396
Number of Sequences: 1393205
Number of extensions: 10634155
Number of successful extensions: 27037
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 26211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27014
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)