Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC004007A_C01 KMC004007A_c01
(793 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
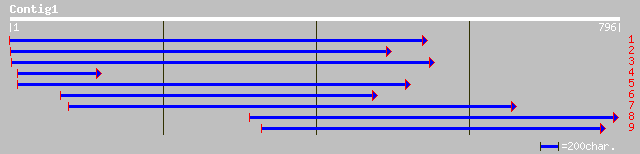
ref|NP_568349.1| root cap 1 (RCP1); protein id: At5g17520.1, sup... 215 7e-55
gb|AAF04351.1|AF168391_1 root cap 1 [Arabidopsis thaliana] gi|21... 215 7e-55
pir||T51478 root cap 1 (RCP1) - Arabidopsis thaliana gi|9755776|... 118 1e-25
ref|NP_484990.1| hypothetical protein [Nostoc sp. PCC 7120] gi|2... 34 2.8
ref|NP_222860.1| putative [Helicobacter pylori J99] gi|7464617|p... 33 3.6
>ref|NP_568349.1| root cap 1 (RCP1); protein id: At5g17520.1, supported by cDNA:
10216., supported by cDNA: gi_15810296, supported by
cDNA: gi_20466028, supported by cDNA: gi_6137137
[Arabidopsis thaliana]
gi|6137138|gb|AAF04350.1|AF168390_1 root cap 1
[Arabidopsis thaliana] gi|15810297|gb|AAL07036.1|
putative root cap protein RCP1 [Arabidopsis thaliana]
gi|20466029|gb|AAM20349.1| putative root cap protein
RCP1 [Arabidopsis thaliana]
Length = 415
Score = 215 bits (547), Expect = 7e-55
Identities = 90/133 (67%), Positives = 117/133 (87%)
Frame = -3
Query: 791 EGVKFVGGISGWTATLLFMWMPVSQMWTNILNPENMKGLSAFSMLLAMLGNGLLLPRALL 612
+GV+FVG +SGWTATL+FMWMPVSQMWTN LNP+N+KGLS+ +MLL+M+GNGL++PRAL
Sbjct: 282 KGVRFVGSLSGWTATLMFMWMPVSQMWTNFLNPDNIKGLSSITMLLSMMGNGLMIPRALF 341
Query: 611 IRDLMWFIGSSWATLFYGYGNLACLFVLNIISKEFFLAATVGLVAWIGMAFWRDSAVHGH 432
IRDLMW GS WATLFYGYGN+ CL+++N S+ FF+AAT+GL++WIG+A WRD+ +GH
Sbjct: 342 IRDLMWLTGSLWATLFYGYGNILCLYLVNCTSQSFFVAATIGLISWIGLALWRDAVAYGH 401
Query: 431 NSPLASIRDLVFG 393
NSP S+++LVFG
Sbjct: 402 NSPFRSLKELVFG 414
>gb|AAF04351.1|AF168391_1 root cap 1 [Arabidopsis thaliana] gi|21536538|gb|AAM60870.1| root
cap 1 (RCP1) [Arabidopsis thaliana]
Length = 415
Score = 215 bits (547), Expect = 7e-55
Identities = 90/133 (67%), Positives = 117/133 (87%)
Frame = -3
Query: 791 EGVKFVGGISGWTATLLFMWMPVSQMWTNILNPENMKGLSAFSMLLAMLGNGLLLPRALL 612
+GV+FVG +SGWTATL+FMWMPVSQMWTN LNP+N+KGLS+ +MLL+M+GNGL++PRAL
Sbjct: 282 KGVRFVGSLSGWTATLMFMWMPVSQMWTNFLNPDNIKGLSSITMLLSMMGNGLMIPRALF 341
Query: 611 IRDLMWFIGSSWATLFYGYGNLACLFVLNIISKEFFLAATVGLVAWIGMAFWRDSAVHGH 432
IRDLMW GS WATLFYGYGN+ CL+++N S+ FF+AAT+GL++WIG+A WRD+ +GH
Sbjct: 342 IRDLMWLTGSLWATLFYGYGNILCLYLVNCTSQSFFVAATIGLISWIGLALWRDAVAYGH 401
Query: 431 NSPLASIRDLVFG 393
NSP S+++LVFG
Sbjct: 402 NSPFRSLKELVFG 414
>pir||T51478 root cap 1 (RCP1) - Arabidopsis thaliana gi|9755776|emb|CAC01896.1|
root cap 1 (RCP1) [Arabidopsis thaliana]
Length = 347
Score = 118 bits (295), Expect = 1e-25
Identities = 50/66 (75%), Positives = 62/66 (93%)
Frame = -3
Query: 791 EGVKFVGGISGWTATLLFMWMPVSQMWTNILNPENMKGLSAFSMLLAMLGNGLLLPRALL 612
+GV+FVG +SGWTATL+FMWMPVSQMWTN LNP+N+KGLS+ +MLL+M+GNGL++PRAL
Sbjct: 282 KGVRFVGSLSGWTATLMFMWMPVSQMWTNFLNPDNIKGLSSITMLLSMMGNGLMIPRALF 341
Query: 611 IRDLMW 594
IRDLMW
Sbjct: 342 IRDLMW 347
>ref|NP_484990.1| hypothetical protein [Nostoc sp. PCC 7120] gi|25354817|pir||AH1924
hypothetical protein alr0947 [imported] - Nostoc sp.
(strain PCC 7120) gi|17130293|dbj|BAB72904.1|
ORF_ID:alr0947~hypothetical protein [Nostoc sp. PCC
7120]
Length = 275
Score = 33.9 bits (76), Expect = 2.8
Identities = 29/108 (26%), Positives = 49/108 (44%), Gaps = 7/108 (6%)
Frame = -3
Query: 743 LFMWMPVSQMWTNILNPENMKGLSAFSMLLAMLGNGLLLPRALLIRDLMWFIGSSWATLF 564
L W+P++ + +LN + +K L L M+ LL P L++ W SS+A
Sbjct: 18 LVCWLPIAAVSLKLLNWQPIKPLQPEQKLPLMISLYLLAP--LVLWGATWLTNSSFAN-- 73
Query: 563 YG-YGNLACL------FVLNIISKEFFLAATVGLVAWIGMAFWRDSAV 441
YG GN A L F L ++S + W+G ++++S +
Sbjct: 74 YGIVGNFALLSSLALGFSLGVLSIVILFTCQL----WLGWCYFKESNI 117
>ref|NP_222860.1| putative [Helicobacter pylori J99] gi|7464617|pir||E71967
hypothetical protein jhp0139 - Helicobacter pylori
(strain J99) gi|4154664|gb|AAD05732.1| putative
[Helicobacter pylori J99]
Length = 255
Score = 33.5 bits (75), Expect = 3.6
Identities = 22/82 (26%), Positives = 40/82 (47%), Gaps = 5/82 (6%)
Frame = -3
Query: 683 KGLSAFSM---LLAMLGNGLLLPRALLIRDLMWFIGSSWATLFYGYGNLACLFVLNIISK 513
+G+ +FS+ L ++L +LLP I L WF+ S + + LF+L+ K
Sbjct: 78 RGVLSFSLPISLESLLLPKILLPMVFFIFSLFWFVASVRLGYYLFNAQSSVLFILHTALK 137
Query: 512 EFFL--AATVGLVAWIGMAFWR 453
F L T+G+ ++G+ +
Sbjct: 138 TFALKPTKTIGVALFLGLVLMK 159
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 689,551,932
Number of Sequences: 1393205
Number of extensions: 15512209
Number of successful extensions: 50009
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 48268
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 49977
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 39775824764
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)