Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
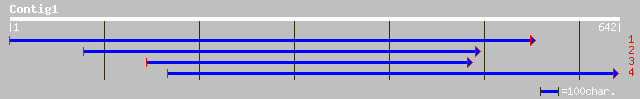
Query= KMC004000A_C01 KMC004000A_c01
(563 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_174469.1| phosphoribosyl-ATP pyrophosphohydrolase (At-IE)... 154 9e-37
dbj|BAA88183.1| unnamed protein product [Oryza sativa (japonica ... 140 1e-32
gb|ZP_00009915.1| hypothetical protein [Rhodopseudomonas palustris] 67 1e-10
gb|ZP_00002423.1| hypothetical protein [Nitrosomonas europaea] 61 1e-08
ref|NP_767295.1| phosphoribosyl-ATP pyrophosphohydrolase [Bradyr... 60 2e-08
>ref|NP_174469.1| phosphoribosyl-ATP pyrophosphohydrolase (At-IE); protein id:
At1g31860.1, supported by cDNA: 25538., supported by
cDNA: gi_15028394, supported by cDNA: gi_19310752,
supported by cDNA: gi_3461883 [Arabidopsis thaliana]
gi|11132859|sp|O82768|HIS2_ARATH Histidine biosynthesis
bifunctional protein hisIE, chloroplast precursor
[Includes: Phosphoribosyl-AMP cyclohydrolase (PRA-CH);
Phosphoribosyl-ATP pyrophosphatase (PRA-PH)]
gi|11358620|pir||T51812 phosphoribosyl-AMP
cyclohydrolase (EC 3.5.4.19) / phosphoribosyl-ATP
diphosphatase (EC 3.6.1.31) [validated] - Arabidopsis
thaliana gi|3461884|dbj|BAA32528.1| phosphoribosyl-ATP
pyrophosphohydrolase [Arabidopsis thaliana]
gi|3461886|dbj|BAA32529.1| phosphoribosyl-ATP
pyrophosphohydrolase [Arabidopsis thaliana]
gi|12321304|gb|AAG50725.1|AC079041_18 phosphoribosyl-ATP
pyrophosphohydrolase (At-IE) [Arabidopsis thaliana]
gi|15028395|gb|AAK76674.1| putative phosphoribosyl-ATP
pyrophosphohydrolase At-IE [Arabidopsis thaliana]
gi|19310753|gb|AAL85107.1| putative phosphoribosyl-ATP
pyrophosphohydrolase At-IE [Arabidopsis thaliana]
gi|21554409|gb|AAM63514.1| phosphoribosyl-ATP
pyrophosphohydrolase At-IE [Arabidopsis thaliana]
Length = 281
Score = 154 bits (388), Expect = 9e-37
Identities = 76/106 (71%), Positives = 92/106 (86%)
Frame = -3
Query: 561 TSLYDLESTISQRKAELAEVENGKPSWTKRLLLNDELLCSKIREEANELCQTLESNEDKS 382
T+LY LES IS+RK E + GKPSWT+RLL +D LLCSKIREEA+ELC+TLE NE+ S
Sbjct: 174 TTLYSLESIISKRKEESTVPQEGKPSWTRRLLTDDALLCSKIREEADELCRTLEDNEEVS 233
Query: 381 RTASEMADVLYHAMVLMARKDVKMEDVLQVLRQRFSQSGIEEKRSR 244
RT SEMADVLYHAMVL++++ VKMEDVL+VLR+RFSQSGIEEK++R
Sbjct: 234 RTPSEMADVLYHAMVLLSKRGVKMEDVLEVLRKRFSQSGIEEKQNR 279
>dbj|BAA88183.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 202
Score = 140 bits (352), Expect = 1e-32
Identities = 71/106 (66%), Positives = 86/106 (80%)
Frame = -3
Query: 561 TSLYDLESTISQRKAELAEVENGKPSWTKRLLLNDELLCSKIREEANELCQTLESNEDKS 382
++LY LE TIS+RK E+ +GKPSWTK+L+L++ LLCSKI EEA EL QTL NED+S
Sbjct: 94 STLYSLEDTISRRKEEIVTEGSGKPSWTKKLILDNRLLCSKISEEAGELNQTLLENEDES 153
Query: 381 RTASEMADVLYHAMVLMARKDVKMEDVLQVLRQRFSQSGIEEKRSR 244
RT SEM D+LYHAMVL+ K V+ME VL+VLR+RFSQSGIEEK SR
Sbjct: 154 RTISEMGDLLYHAMVLLRVKGVRMEQVLEVLRKRFSQSGIEEKASR 199
>gb|ZP_00009915.1| hypothetical protein [Rhodopseudomonas palustris]
Length = 107
Score = 67.4 bits (163), Expect = 1e-10
Identities = 44/106 (41%), Positives = 61/106 (57%), Gaps = 1/106 (0%)
Frame = -3
Query: 558 SLYDLESTISQRKAELAEVENGKPSWTKRLLLNDELLCSK-IREEANELCQTLESNEDKS 382
+L+DL +T+ R A G+ S+TK+LL C+K EEA E+ N D+
Sbjct: 5 TLHDLAATVDARAAS-----GGESSYTKKLLDKGPEHCAKKFGEEAVEMVIAAVEN-DRG 58
Query: 381 RTASEMADVLYHAMVLMARKDVKMEDVLQVLRQRFSQSGIEEKRSR 244
SE ADVL+H +VL+ + VK+E+V L QR S SG+EEK SR
Sbjct: 59 HLISETADVLFHMLVLLKSRGVKLEEVEAALAQRTSMSGLEEKASR 104
>gb|ZP_00002423.1| hypothetical protein [Nitrosomonas europaea]
Length = 112
Score = 60.8 bits (146), Expect = 1e-08
Identities = 40/110 (36%), Positives = 61/110 (55%), Gaps = 4/110 (3%)
Frame = -3
Query: 540 STISQRKAELAEV-ENGKPS--WTKRLL-LNDELLCSKIREEANELCQTLESNEDKSRTA 373
STI QR A E +N PS +T +LL + + + KI EEA E + N D+ +
Sbjct: 4 STILQRLARTIEARKNADPSISYTAKLLNSSQDKVLKKIAEEAAETIMACKDN-DREQII 62
Query: 372 SEMADVLYHAMVLMARKDVKMEDVLQVLRQRFSQSGIEEKRSRASQKSVD 223
E AD+ +H ++++ R D+ ED+L+ L +R SGIEEK SR+ +
Sbjct: 63 YETADLWFHCLIMLTRHDISPEDILRELERREGISGIEEKLSRSQPNKTE 112
>ref|NP_767295.1| phosphoribosyl-ATP pyrophosphohydrolase [Bradyrhizobium japonicum]
gi|27348904|dbj|BAC45920.1| phosphoribosyl-ATP
pyrophosphohydrolase [Bradyrhizobium japonicum USDA 110]
Length = 106
Score = 59.7 bits (143), Expect = 2e-08
Identities = 40/106 (37%), Positives = 60/106 (55%), Gaps = 1/106 (0%)
Frame = -3
Query: 558 SLYDLESTISQRKAELAEVENGKPSWTKRLLLNDELLCSK-IREEANELCQTLESNEDKS 382
+++DL TI R A G+ S+T++LL C+K EEA E N D++
Sbjct: 5 TIHDLAETIDARAAA-----GGEGSYTRKLLDKGAEHCAKKFGEEAVETVIAAVEN-DRA 58
Query: 381 RTASEMADVLYHAMVLMARKDVKMEDVLQVLRQRFSQSGIEEKRSR 244
+E AD+LYH +VL+ + +K+E+V L +R S SG+EEK SR
Sbjct: 59 HLIAEGADLLYHFLVLLKARGIKLEEVEAALGKRTSMSGLEEKASR 104
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 451,089,931
Number of Sequences: 1393205
Number of extensions: 9178630
Number of successful extensions: 25394
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 24640
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25387
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)