Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003989A_C01 KMC003989A_c01
(597 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
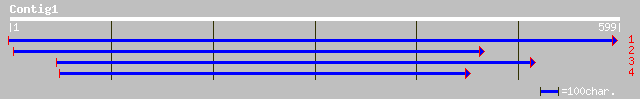
gb|AAA24215.1| glycosyl transferase 33 2.1
pir||A42718 polysialyltransferase - Escherichia coli (strain K92) 33 2.1
ref|NP_187419.1| Exostosin family; protein id: At3g07620.1 [Arab... 33 2.1
ref|NP_014552.1| involved in salt tolerance; Hal9p [Saccharomyce... 33 2.8
pir||A61542 poly-alpha-2,8 sialosyl sialyltransferase (EC 2.4.-.... 32 4.7
>gb|AAA24215.1| glycosyl transferase
Length = 409
Score = 33.5 bits (75), Expect = 2.1
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Frame = +3
Query: 327 QLNKIW-NSCAFPLPSSFMNPEPKLYELLFKW*PRKPKSKTIFIKDELGLINKEKLELEF 503
+L K++ N F S F N + K ELL P K KSK IFI LG + K +L ++
Sbjct: 9 KLRKLFVNPIGFFRDSWFFNSKNKAEELLS---PLKIKSKNIFIVAHLGQLKKAELFIQK 65
Query: 504 FSCKS 518
FS +S
Sbjct: 66 FSRRS 70
>pir||A42718 polysialyltransferase - Escherichia coli (strain K92)
Length = 409
Score = 33.5 bits (75), Expect = 2.1
Identities = 26/65 (40%), Positives = 35/65 (53%), Gaps = 1/65 (1%)
Frame = +3
Query: 327 QLNKIW-NSCAFPLPSSFMNPEPKLYELLFKW*PRKPKSKTIFIKDELGLINKEKLELEF 503
+L K++ N F S F N + K ELL P K KSK IFI LG + K +L ++
Sbjct: 9 KLRKLFVNPIGFFRDSWFFNSKNKAEELLS---PLKIKSKNIFIVAHLGQLKKAELFIQK 65
Query: 504 FSCKS 518
FS +S
Sbjct: 66 FSRRS 70
>ref|NP_187419.1| Exostosin family; protein id: At3g07620.1 [Arabidopsis thaliana]
gi|6466945|gb|AAF13080.1|AC009176_7 hypothetical protein
[Arabidopsis thaliana]
Length = 470
Score = 33.5 bits (75), Expect = 2.1
Identities = 23/64 (35%), Positives = 31/64 (47%), Gaps = 11/64 (17%)
Frame = -3
Query: 505 KNSNSSFSLFISPSSSLINIVFDLGFLGYHL-----------KSNSYNLGSGFMNDEGKG 359
K+SNSS L+ S SSSL F F+ + K N N GSG+ +GK
Sbjct: 29 KDSNSSSHLYFSTSSSLWTSSFSPAFITVSIFLTVHRFREKRKRNGSNPGSGYWKRDGKV 88
Query: 358 KAQL 347
+A+L
Sbjct: 89 EAEL 92
>ref|NP_014552.1| involved in salt tolerance; Hal9p [Saccharomyces cerevisiae]
gi|1362355|pir||S57380 probable membrane protein YOL089c
- yeast (Saccharomyces cerevisiae)
gi|600469|emb|CAA58190.1| orf 00938 [Saccharomyces
cerevisiae] gi|1419932|emb|CAA99101.1| ORF YOL089c
[Saccharomyces cerevisiae]
Length = 1030
Score = 33.1 bits (74), Expect = 2.8
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = +2
Query: 92 HHLVQTNSKNTANNMSKNTSKQHVPTEV*LEKFNG-VPKEVNILEKLANLE 241
HH S T+N++S + VP E NG P E NI++K++N++
Sbjct: 185 HHATPGESLQTSNSISNPVASSSVPNSGRFELLNGNSPLESNIIDKVSNIQ 235
>pir||A61542 poly-alpha-2,8 sialosyl sialyltransferase (EC 2.4.-.-) -
Escherichia coli gi|42116|emb|CAA43053.1|
polysialyltransferase [Escherichia coli]
gi|146949|gb|AAA24213.1| glycosyl transferase
Length = 409
Score = 32.3 bits (72), Expect = 4.7
Identities = 30/89 (33%), Positives = 41/89 (45%), Gaps = 1/89 (1%)
Frame = +3
Query: 327 QLNKIW-NSCAFPLPSSFMNPEPKLYELLFKW*PRKPKSKTIFIKDELGLINKEKLELEF 503
+L K++ N F S F N + K ELL P K KSK IFI LG + K + ++
Sbjct: 9 KLRKLFVNPIGFFRDSWFFNSKNKAEELLS---PLKIKSKNIFIISNLGQLKKAESFVQK 65
Query: 504 FSCKSGTSPSTTTFQLRAGPGMTRRSINN 590
FS +S T + P + INN
Sbjct: 66 FSKRSNYLIVLATEKNTEMPKIIVEQINN 94
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,154,655
Number of Sequences: 1393205
Number of extensions: 11781029
Number of successful extensions: 29735
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 28706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29725
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)