Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003985A_C01 KMC003985A_c01
(801 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
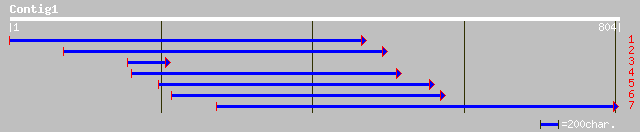
ref|NP_175245.1| unknown protein; protein id: At1g48110.1 [Arabi... 35 1.3
gb|AAF79522.1|AC023673_10 F21D18.17 [Arabidopsis thaliana] 35 1.3
gb|EAA17233.1| hypothetical protein [Plasmodium yoelii yoelii] 35 1.7
gb|AAO51998.1| similar to Leishmania major. Ppg3 [Dictyostelium ... 33 3.7
emb|CAD53105.1| hypothetical protein, unlikely [Trypanosoma brucei] 33 4.9
>ref|NP_175245.1| unknown protein; protein id: At1g48110.1 [Arabidopsis thaliana]
Length = 639
Score = 35.0 bits (79), Expect = 1.3
Identities = 29/88 (32%), Positives = 41/88 (45%), Gaps = 19/88 (21%)
Frame = -3
Query: 799 EDISSVLK-IGSVTITPK----------------QTETKPYSNGVANKEPNDVVTVGSMQ 671
ED SS K I S+TI P Q ++KP S+G K V S+
Sbjct: 551 EDTSSTQKKISSLTIDPSGTDSNPTTVSHLNQKSQAKSKPNSSGSLKKTDPSEVVDASLS 610
Query: 670 VRVNGF--SESSGFLKVGSIPLDPRALK 593
+ + F + S L VG+IPLDP++L+
Sbjct: 611 DKNDSFKVTGSPAILTVGTIPLDPKSLQ 638
>gb|AAF79522.1|AC023673_10 F21D18.17 [Arabidopsis thaliana]
Length = 664
Score = 35.0 bits (79), Expect = 1.3
Identities = 29/88 (32%), Positives = 41/88 (45%), Gaps = 19/88 (21%)
Frame = -3
Query: 799 EDISSVLK-IGSVTITPK----------------QTETKPYSNGVANKEPNDVVTVGSMQ 671
ED SS K I S+TI P Q ++KP S+G K V S+
Sbjct: 576 EDTSSTQKKISSLTIDPSGTDSNPTTVSHLNQKSQAKSKPNSSGSLKKTDPSEVVDASLS 635
Query: 670 VRVNGF--SESSGFLKVGSIPLDPRALK 593
+ + F + S L VG+IPLDP++L+
Sbjct: 636 DKNDSFKVTGSPAILTVGTIPLDPKSLQ 663
>gb|EAA17233.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 205
Score = 34.7 bits (78), Expect = 1.7
Identities = 18/49 (36%), Positives = 26/49 (52%)
Frame = +1
Query: 352 YTNHFNLFGINTSPEIHPQGMLYLINENSKRRLKETQLPNGNKIRQISV 498
YTNHF LF N S EI Q + I+ + K++ K+ L N R + +
Sbjct: 77 YTNHFQLFEYNKSEEIDKQKYINSISRSIKKQKKKMYLLEKNACRILDI 125
>gb|AAO51998.1| similar to Leishmania major. Ppg3 [Dictyostelium discoideum]
Length = 642
Score = 33.5 bits (75), Expect = 3.7
Identities = 26/84 (30%), Positives = 36/84 (41%), Gaps = 3/84 (3%)
Frame = +3
Query: 486 ADISPSFTREDPNSALTSISKF*GLDPFLTRVPLS---GFKALGSSGMLPTFKKPEDSLN 656
+D PSF P+ + ++S F G P + PLS G SSG P+ D +
Sbjct: 350 SDSLPSFPGSFPSGSSDTLSSFPGSLPSDSSDPLSSSPGSLPSDSSGSFPSLTGSSDLSS 409
Query: 657 PLTLTCIEPTVTTSFGSLLATPLL 728
+T T PT T +F P L
Sbjct: 410 TITTTPTTPTTTPNFTFTPTIPTL 433
>emb|CAD53105.1| hypothetical protein, unlikely [Trypanosoma brucei]
Length = 99
Score = 33.1 bits (74), Expect = 4.9
Identities = 18/35 (51%), Positives = 25/35 (71%), Gaps = 2/35 (5%)
Frame = -1
Query: 261 LPYSTIVL-CNSAMFFPIFFLFYLLECV-LV*FVF 163
+P+ TI+L C FFP+FFLF+L E + +V FVF
Sbjct: 49 IPHLTILLLCCCCCFFPLFFLFFLGEAINVVLFVF 83
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 665,457,681
Number of Sequences: 1393205
Number of extensions: 14382575
Number of successful extensions: 32853
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 31603
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32841
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40616159090
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)