Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003976A_C01 KMC003976A_c01
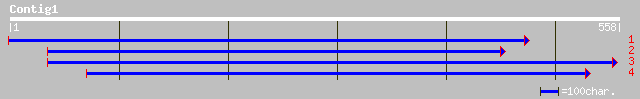
(548 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC42055.1| unknown protein [Arabidopsis thaliana] 113 2e-24
gb|AAO72596.1| unknown [Oryza sativa (japonica cultivar-group)] 58 7e-08
gb|AAL83628.1|AC093017_12 putative growth factor [Oryza sativa (... 49 3e-05
ref|NP_175098.1| unknown protein; protein id: At1g44770.1 [Arabi... 43 0.002
pir||F96506 hypothetical protein T12C22.4 [imported] - Arabidops... 42 0.005
>dbj|BAC42055.1| unknown protein [Arabidopsis thaliana]
Length = 241
Score = 113 bits (282), Expect = 2e-24
Identities = 52/75 (69%), Positives = 65/75 (86%)
Frame = -2
Query: 499 QSRTLTPNQLQEALSQTFSLKKKKGKLRKAWDGSQVIYNVASWGATAIGIYPNPVILRAA 320
Q++ ++P+QLQ+ALS FS+KK+KGKLRKAW+GS+VIYNVASW ATAIGIY NP+IL A
Sbjct: 167 QTKDISPDQLQKALSTMFSVKKRKGKLRKAWEGSKVIYNVASWSATAIGIYQNPMILSIA 226
Query: 319 TKAFWTSCHVISKLL 275
+KAFW SC ISKL+
Sbjct: 227 SKAFWMSCQAISKLV 241
Score = 38.1 bits (87), Expect = 0.070
Identities = 21/51 (41%), Positives = 26/51 (50%)
Frame = -1
Query: 545 EGIACFMAAYLLSLQPIQDFDPQSTPGSSQPDILSEKEKGKASKGMGWEPS 393
+GIACFMA YL SL+ +D P + +K KGK K WE S
Sbjct: 152 DGIACFMATYLSSLKQTKDISPDQLQKALSTMFSVKKRKGKLRK--AWEGS 200
>gb|AAO72596.1| unknown [Oryza sativa (japonica cultivar-group)]
Length = 377
Score = 58.2 bits (139), Expect = 7e-08
Identities = 25/43 (58%), Positives = 34/43 (78%)
Frame = -2
Query: 499 QSRTLTPNQLQEALSQTFSLKKKKGKLRKAWDGSQVIYNVASW 371
Q++ L PNQLQ+ALS+TF KK+K L+K DG++VIYN+ SW
Sbjct: 247 QTKDLXPNQLQQALSKTFXAKKRKXXLQKXXDGTKVIYNIXSW 289
Score = 49.7 bits (117), Expect = 2e-05
Identities = 31/94 (32%), Positives = 43/94 (44%)
Frame = -1
Query: 548 VEGIACFMAAYLLSLQPIQDFDPQSTPGSSQPDILSEKEKGKASKGMGWEPSYL*CCFMG 369
V+GIA MAAYLLSL+ +D P + ++K K K
Sbjct: 231 VDGIASXMAAYLLSLKQTKDLXPNQLQQALSKTFXAKKRKXXLQKXXDGTKVIYNIXSWE 290
Query: 368 GYSYWDISKPCYSKGSNQSILDFLPCYIKATLIS 267
YW + + SK SN +LDFLPC I+ ++ S
Sbjct: 291 CNCYWHLPESSNSKSSNCCLLDFLPCSIQVSVNS 324
>gb|AAL83628.1|AC093017_12 putative growth factor [Oryza sativa (japonica cultivar-group)]
Length = 479
Score = 49.3 bits (116), Expect = 3e-05
Identities = 20/62 (32%), Positives = 39/62 (62%)
Frame = -2
Query: 499 QSRTLTPNQLQEALSQTFSLKKKKGKLRKAWDGSQVIYNVASWGATAIGIYPNPVILRAA 320
+S++++P +LQ+A++ S K K W+ +V+Y +A+WG T +G+Y + +L+ A
Sbjct: 184 KSKSISPVELQKAVAMALSTLNDKWKWMSIWEAGKVLYILATWGITIVGLYRSRHVLKIA 243
Query: 319 TK 314
K
Sbjct: 244 AK 245
>ref|NP_175098.1| unknown protein; protein id: At1g44770.1 [Arabidopsis thaliana]
gi|27808560|gb|AAO24560.1| At1g44770 [Arabidopsis
thaliana]
Length = 271
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/75 (26%), Positives = 38/75 (50%)
Frame = -2
Query: 499 QSRTLTPNQLQEALSQTFSLKKKKGKLRKAWDGSQVIYNVASWGATAIGIYPNPVILRAA 320
+++ L P +LQ+ L Q + K GK+ W ++ Y +++WG G+Y +L+ A
Sbjct: 197 KAKNLKPEELQKLLVQEVTALSKVGKVVDIWHAGKMFYTLSTWGLAFGGLYQARGVLKIA 256
Query: 319 TKAFWTSCHVISKLL 275
K + V+ + L
Sbjct: 257 AKGVHATSKVVLRAL 271
>pir||F96506 hypothetical protein T12C22.4 [imported] - Arabidopsis thaliana
gi|8655987|gb|AAF78260.1|AC020576_4 Contains weak
similarity to tail completion gi|5354213 from coliphage
T4 gb|AF158101. ESTs gb|AA650799, gb|AA041054,
gb|R29873, gb|AA712908 come from this gene. [Arabidopsis
thaliana]
Length = 269
Score = 42.0 bits (97), Expect = 0.005
Identities = 20/73 (27%), Positives = 36/73 (48%)
Frame = -2
Query: 493 RTLTPNQLQEALSQTFSLKKKKGKLRKAWDGSQVIYNVASWGATAIGIYPNPVILRAATK 314
+ L P +LQ+ L Q + K GK+ W ++ Y +++WG G+Y +L+ A K
Sbjct: 197 QNLKPEELQKLLVQEVTALSKVGKVVDIWHAGKMFYTLSTWGLAFGGLYQARGVLKIAAK 256
Query: 313 AFWTSCHVISKLL 275
+ V+ + L
Sbjct: 257 GVHATSKVVLRAL 269
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,549,231
Number of Sequences: 1393205
Number of extensions: 10412326
Number of successful extensions: 28667
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 27723
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28600
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)