Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003973A_C01 KMC003973A_c01
(650 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
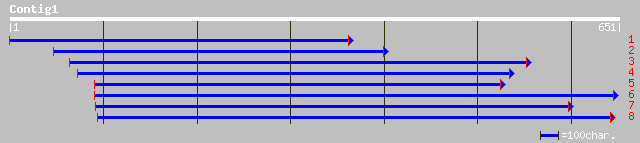
ref|NP_175685.1| putative lipase; protein id: At1g52760.1, suppo... 267 1e-70
ref|NP_172576.1| lysophospholipase isolog; protein id: At1g11090... 122 4e-27
dbj|BAC22550.1| putative lysophospholipase isolog [Oryza sativa ... 122 4e-27
ref|NP_191079.1| lipase -like protein; protein id: At3g55190.1 [... 118 6e-26
dbj|BAB90342.1| phospholipase-like protein [Oryza sativa (japoni... 116 2e-25
>ref|NP_175685.1| putative lipase; protein id: At1g52760.1, supported by cDNA:
36954., supported by cDNA: gi_15450993 [Arabidopsis
thaliana] gi|25405597|pir||F96568 probable lipase,
20450-21648 [imported] - Arabidopsis thaliana
gi|12324637|gb|AAG52273.1|AC019018_10 putative lipase;
20450-21648 [Arabidopsis thaliana]
gi|15450994|gb|AAK96768.1| putative lipase [Arabidopsis
thaliana]
Length = 332
Score = 267 bits (682), Expect = 1e-70
Identities = 124/153 (81%), Positives = 140/153 (91%)
Frame = -2
Query: 649 PEDMKPSRVHLFLYGLLFGVADTWAAMPDNKIVGKAIRDPQKLKVIASNPRRYTGPPRVG 470
PEDMKPS+ HLF YGLLFG+ADTWAAMPDNK+VGKAI+DP+KLK+IASNP+RYTG PRVG
Sbjct: 178 PEDMKPSKAHLFAYGLLFGLADTWAAMPDNKMVGKAIKDPEKLKIIASNPQRYTGKPRVG 237
Query: 469 TMRELLRVTQYVQDNFSKVTTPFLTAHGTADGVTCPSSSKLLYEKASCEDKTLKLYDGMY 290
TMRELLR TQYVQ+NF KVT P TAHGTADGVTCP+SSKLLYEKAS DKTLK+Y+GMY
Sbjct: 238 TMRELLRKTQYVQENFGKVTIPVFTAHGTADGVTCPTSSKLLYEKASSADKTLKIYEGMY 297
Query: 289 HSLIQGEPDESANLVLGDMREWIDERVRRYGPK 191
HSLIQGEPDE+A +VL DMREWIDE+V++YG K
Sbjct: 298 HSLIQGEPDENAEIVLKDMREWIDEKVKKYGSK 330
>ref|NP_172576.1| lysophospholipase isolog; protein id: At1g11090.1 [Arabidopsis
thaliana] gi|25402643|pir||H86244 lysophospholipase
homolog, 25331-24357 [imported] - Arabidopsis thaliana
gi|1931639|gb|AAB65474.1| lysophospholipase isolog;
25331-24357 [Arabidopsis thaliana]
gi|26450507|dbj|BAC42367.1| putative lysophospholipase
isolog [Arabidopsis thaliana]
Length = 324
Score = 122 bits (306), Expect = 4e-27
Identities = 56/124 (45%), Positives = 81/124 (65%)
Frame = -2
Query: 583 TWAAMPDNKIVGKAIRDPQKLKVIASNPRRYTGPPRVGTMRELLRVTQYVQDNFSKVTTP 404
TWA +P ++ K+I+ +K + NP RY PR+GT+ ELLRVT Y+ V+ P
Sbjct: 191 TWAIVPTEDLLEKSIKVEEKKPIAKRNPMRYNEKPRLGTVMELLRVTDYLGKKLKDVSIP 250
Query: 403 FLTAHGTADGVTCPSSSKLLYEKASCEDKTLKLYDGMYHSLIQGEPDESANLVLGDMREW 224
F+ HG+AD VT P S+ LYE A +DKTLK+YDGM HS++ GEPD++ +V D+ W
Sbjct: 251 FIIVHGSADAVTDPEVSRELYEHAKSKDKTLKIYDGMMHSMLFGEPDDNIEIVRKDIVSW 310
Query: 223 IDER 212
+++R
Sbjct: 311 LNDR 314
>dbj|BAC22550.1| putative lysophospholipase isolog [Oryza sativa (japonica
cultivar-group)]
Length = 334
Score = 122 bits (306), Expect = 4e-27
Identities = 58/126 (46%), Positives = 83/126 (65%)
Frame = -2
Query: 589 ADTWAAMPDNKIVGKAIRDPQKLKVIASNPRRYTGPPRVGTMRELLRVTQYVQDNFSKVT 410
A T A +P ++ K+++ P K + A NP RY+G PR+GT+ ELLR T + +VT
Sbjct: 195 APTLAIVPTADLIEKSVKVPAKRLIAARNPMRYSGRPRLGTVVELLRATDELGARLGEVT 254
Query: 409 TPFLTAHGTADGVTCPSSSKLLYEKASCEDKTLKLYDGMYHSLIQGEPDESANLVLGDMR 230
PFL HG+AD VT P S+ LY+ A+ +DKT+K+YDGM HS++ GEPDE+ V D+
Sbjct: 255 VPFLVVHGSADEVTDPDISRALYDAAASKDKTIKIYDGMMHSMLFGEPDENIERVRADIL 314
Query: 229 EWIDER 212
W++ER
Sbjct: 315 AWLNER 320
>ref|NP_191079.1| lipase -like protein; protein id: At3g55190.1 [Arabidopsis
thaliana] gi|11358487|pir||T47658 lipase-like protein -
Arabidopsis thaliana gi|7019652|emb|CAB75753.1|
lipase-like protein [Arabidopsis thaliana]
Length = 319
Score = 118 bits (296), Expect = 6e-26
Identities = 63/158 (39%), Positives = 96/158 (59%), Gaps = 3/158 (1%)
Frame = -2
Query: 646 EDMKPSRVHLFLYGLLFGVADTWAAM---PDNKIVGKAIRDPQKLKVIASNPRRYTGPPR 476
E+MKPSR+ + + ++ + +W ++ PD I+ AI+ P+K I +NP Y G PR
Sbjct: 150 EEMKPSRMVISMINMVTNLIPSWKSIIHGPD--ILNSAIKLPEKRHEIRTNPNCYNGWPR 207
Query: 475 VGTMRELLRVTQYVQDNFSKVTTPFLTAHGTADGVTCPSSSKLLYEKASCEDKTLKLYDG 296
+ TM EL R++ +++ ++VT PF+ HG D VT SKLLYE A DKTLKLY
Sbjct: 208 MKTMSELFRISLDLENRLNEVTMPFIVLHGEDDKVTDKGGSKLLYEVALSNDKTLKLYPE 267
Query: 295 MYHSLIQGEPDESANLVLGDMREWIDERVRRYGPKNNS 182
M+HSL+ GEP E++ +V D+ +W+ R+ K N+
Sbjct: 268 MWHSLLFGEPPENSEIVFNDIVQWMQTRITTLQVKANN 305
>dbj|BAB90342.1| phospholipase-like protein [Oryza sativa (japonica cultivar-group)]
Length = 349
Score = 116 bits (291), Expect = 2e-25
Identities = 58/149 (38%), Positives = 83/149 (54%)
Frame = -2
Query: 646 EDMKPSRVHLFLYGLLFGVADTWAAMPDNKIVGKAIRDPQKLKVIASNPRRYTGPPRVGT 467
E +KP V + L + V W +P ++ A +DP K + I N Y PR+ T
Sbjct: 145 EKVKPHPVVISLLTQVEDVIPRWKIVPTKDVIDAAFKDPAKREKIRKNKLIYQDKPRLKT 204
Query: 466 MRELLRVTQYVQDNFSKVTTPFLTAHGTADGVTCPSSSKLLYEKASCEDKTLKLYDGMYH 287
E+LR + YV+D+ SKV PF HG AD VT P S+ LYE+A+ DK +KLY GM+H
Sbjct: 205 ALEMLRTSMYVEDSLSKVKLPFFVLHGDADTVTDPEVSRALYERAASADKAIKLYAGMWH 264
Query: 286 SLIQGEPDESANLVLGDMREWIDERVRRY 200
L GEPD + + + D+ W++ R R +
Sbjct: 265 GLTAGEPDHNVDAIFSDIVAWLNGRSRTW 293
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 606,102,495
Number of Sequences: 1393205
Number of extensions: 14549811
Number of successful extensions: 42391
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 39489
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42247
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)