Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003970A_C01 KMC003970A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
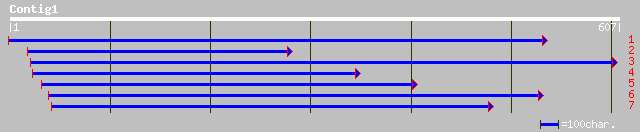
sp|Q01443|SSP2_PLAYO Sporozoite surface protein 2 precursor gi|3... 41 0.013
gb|AAO50991.1| hypothetical protein [Dictyostelium discoideum] 39 0.048
gb|AAO52430.1| similar to Dictyostelium discoideum (Slime mold).... 39 0.063
gb|AAO51825.1| similar to Dictyostelium discoideum (Slime mold).... 38 0.082
emb|CAC38114.1| KIAA0298 protein [Mus musculus] 38 0.082
>sp|Q01443|SSP2_PLAYO Sporozoite surface protein 2 precursor gi|323142|pir||A45559
sporozoite surface protein 2 - Plasmodium yoelii
gi|160693|gb|AAA29768.1| sporozoite surface protein
Length = 826
Score = 40.8 bits (94), Expect = 0.013
Identities = 41/152 (26%), Positives = 56/152 (35%), Gaps = 13/152 (8%)
Frame = +2
Query: 167 NPNKALNNQRARSIRRGPRCGEKKPRKPNKNKTSSRHWGNTN*HNTSTIGS--------- 319
NPN + +R RR P + KP KPN NK + N N N + +
Sbjct: 386 NPNN--HPKRRNPKRRNPN--KPKPNKPNPNKPNPNEPSNPNKPNPNEPSNPNKPNPNEP 441
Query: 320 ---NKTEPNFQVNHGLLQNSNSVGPAHQTTKLERSQ*QCNLDISSAKPTTYRTNANEP-N 487
NK PN N + + P + E S + + +N NEP N
Sbjct: 442 SNPNKPNPNEPSNPNKPNPNEPLNPNEPSNPNEPSNPNAPSNPNEPSNPNEPSNPNEPSN 501
Query: 488 SEPPATGQHTKNPKTERPKLSKEPSYAS*PLN 583
P+ NPK +P EPS + PLN
Sbjct: 502 PNEPSNPNEPSNPK--KPSNPNEPSNPNEPLN 531
Score = 33.1 bits (74), Expect = 2.6
Identities = 42/160 (26%), Positives = 55/160 (34%), Gaps = 21/160 (13%)
Frame = +2
Query: 167 NPNKALNNQRARSIRRGPRCGEKKPRKPNKNKT-SSRHWGNTN*HNTSTIGSNKTEP--- 334
NPNK N+ + + P P KPN N+ + N N + SN EP
Sbjct: 432 NPNKPNPNEPSNPNKPNPN-EPSNPNKPNPNEPLNPNEPSNPNEPSNPNAPSNPNEPSNP 490
Query: 335 ------NFQVNHGLLQNSNSVG-PAHQTTKLERSQ*QCNLDISSAKPTTYRTNANEP-NS 490
N N N N P + E S L+ + +N NEP N
Sbjct: 491 NEPSNPNEPSNPNEPSNPNEPSNPKKPSNPNEPSNPNEPLNPNEPSNPNEPSNPNEPSNP 550
Query: 491 EPPATGQHTKNPKT---------ERPKLSKEPSYAS*PLN 583
E P+ + NP E P KEPS P+N
Sbjct: 551 EEPSNPKEPSNPNEPSNPEEPNPEEPSNPKEPSNPEEPIN 590
>gb|AAO50991.1| hypothetical protein [Dictyostelium discoideum]
Length = 1483
Score = 38.9 bits (89), Expect = 0.048
Identities = 32/157 (20%), Positives = 61/157 (38%), Gaps = 2/157 (1%)
Frame = +2
Query: 68 RVPQIVQSKPKNEFIP*NFTKQQTNLEPQP*GLNPNKALNNQRARSIRRGPRCGEKKPRK 247
++ Q +Q + + + +T QQT+ P ++ K + + P +
Sbjct: 870 QIQQQLQQQQQQQQQQQQWTSQQTHTNTTPNMVS--KISTTPQTSNNSNKPTYAPPPQFQ 927
Query: 248 PNKNKTSSRHWGNTN*HNTSTIGSNKTEPNFQVNHGLLQNSNSVGPAHQTTKLERSQ*QC 427
N N ++ + N N +N + +N N +N+ N+ S+ P + +
Sbjct: 928 TNNNNNNNNNNNNNNNNNNNNNNNNNNNSNTNINNN--NNNTSMNPPTPNSSTSMNPPTP 985
Query: 428 NLDISSAKPTTYRTNANEPNSEP--PATGQHTKNPKT 532
N ++ P T T+ N P P P+T NP T
Sbjct: 986 NTSMNPPTPNTVNTSMNPPTPTPATPSTPSTMMNPPT 1022
>gb|AAO52430.1| similar to Dictyostelium discoideum (Slime mold).
Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 2084
Score = 38.5 bits (88), Expect = 0.063
Identities = 33/136 (24%), Positives = 56/136 (40%), Gaps = 3/136 (2%)
Frame = +2
Query: 173 NKALNNQRARSIRRGPR---CGEKKPRKPNKNKTSSRHWGNTN*HNTSTIGSNKTEPNFQ 343
N AL A +I + PR N N ++ + N N +N +TI + +T Q
Sbjct: 1823 NIALTTSNA-TISQSPRQSTVNNNNNNNNNNNNNNNNNNNNNNNNNNNTIITPRTSFQQQ 1881
Query: 344 VNHGLLQNSNSVGPAHQTTKLERSQ*QCNLDISSAKPTTYRTNANEPNSEPPATGQHTKN 523
+ L NS + P+ Q K+ S N + S +P T+++ +S ++
Sbjct: 1882 QQNASLANSTTTAPSVQKFKINTSTIPTNNGLVSPRPPLPTTSSSSSSSSSSSSATINNG 1941
Query: 524 PKTERPKLSKEPSYAS 571
+ RP L S +S
Sbjct: 1942 LVSPRPPLPTSSSNSS 1957
>gb|AAO51825.1| similar to Dictyostelium discoideum (Slime mold). Coronin binding
protein
Length = 507
Score = 38.1 bits (87), Expect = 0.082
Identities = 28/122 (22%), Positives = 42/122 (33%)
Frame = +2
Query: 167 NPNKALNNQRARSIRRGPRCGEKKPRKPNKNKTSSRHWGNTN*HNTSTIGSNKTEPNFQV 346
N N NN R+I G + N N + N+N +N S SN N
Sbjct: 295 NSNNNSNNSNNRNITNGSNANKSNSPNNNLNTNNDNKNNNSNNNNNSNNNSNNGNSNNNN 354
Query: 347 NHGLLQNSNSVGPAHQTTKLERSQ*QCNLDISSAKPTTYRTNANEPNSEPPATGQHTKNP 526
N+ ++ N+NS N + +S + +N N PN + N
Sbjct: 355 NNNIINNNNS---------------NSNSNNNSNNNSNNNSNRNSPNHNNNGDNDNNTNN 399
Query: 527 KT 532
T
Sbjct: 400 NT 401
>emb|CAC38114.1| KIAA0298 protein [Mus musculus]
Length = 1209
Score = 38.1 bits (87), Expect = 0.082
Identities = 42/166 (25%), Positives = 70/166 (41%), Gaps = 14/166 (8%)
Frame = +1
Query: 109 HPIKLYKATNKPR------ATTLRPKPE*SIKQPTSKKYPSGT*MRREKASQTQQKQNLI 270
HP KL + +P+ +T+ RP P+ + QP P A T Q ++
Sbjct: 378 HPPKLMQPWLEPQPPAEQESTSQRPGPQ-LVSQPVCIVPPQDV-QPGAHAQPTIQTPSIQ 435
Query: 271 SALGEHELAQHINHRQQQNRTKFPS*PWPPPKL*FGWTGTPDNEIRAKSMTM*PRHKQCE 450
LG H+ + ++H QQQ + + P P PPP P ++ A S P C
Sbjct: 436 VQLGHHQKLK-LSHFQQQPQQQPPPPPPPPPPPQHAPPPLPPSQHLASSQHESPPGPACS 494
Query: 451 -----THH---IQNQRQRTQLRTSCDRPTHQKPQNRAPKTKQGTIL 564
HH ++ ++ +L +P+ Q Q ++P+ Q TI+
Sbjct: 495 QNVDIMHHKFELEEMQKDLELLLQAQQPSLQLSQTKSPQHLQQTIV 540
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 549,515,113
Number of Sequences: 1393205
Number of extensions: 12952056
Number of successful extensions: 49853
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 39710
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46550
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)