Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003956A_C01 KMC003956A_c01
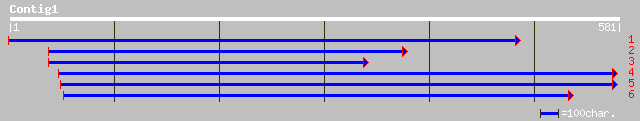
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_181448.1| unknown protein; protein id: At2g39150.1 [Arabi... 79 4e-14
gb|AAM66944.1| unknown [Arabidopsis thaliana] gi|26450440|dbj|BA... 79 4e-14
ref|NP_189920.1| putative protein; protein id: At3g43340.1 [Arab... 50 3e-05
ref|NP_345185.1| ribosomal small subunit pseudouridine synthase ... 38 0.10
gb|ZP_00035279.1| hypothetical protein [Enterococcus faecium] 37 0.14
>ref|NP_181448.1| unknown protein; protein id: At2g39150.1 [Arabidopsis thaliana]
gi|7487058|pir||T02587 hypothetical protein At2g39150
[imported] - Arabidopsis thaliana
gi|3402691|gb|AAC28994.1| unknown protein [Arabidopsis
thaliana]
Length = 86
Score = 79.0 bits (193), Expect = 4e-14
Identities = 36/50 (72%), Positives = 44/50 (88%)
Frame = -3
Query: 575 EVRELVKRAGLEIYSLKRVRIGGFKLPPDLGIGKYLELNPTHLNALGWKS 426
EVRELVK AGLE++SLKRVRIGGF+LP DLG+GK++EL + L ALGWK+
Sbjct: 37 EVRELVKNAGLEVHSLKRVRIGGFRLPSDLGLGKHVELKQSELKALGWKN 86
>gb|AAM66944.1| unknown [Arabidopsis thaliana] gi|26450440|dbj|BAC42334.1| unknown
protein [Arabidopsis thaliana]
Length = 410
Score = 79.0 bits (193), Expect = 4e-14
Identities = 36/50 (72%), Positives = 44/50 (88%)
Frame = -3
Query: 575 EVRELVKRAGLEIYSLKRVRIGGFKLPPDLGIGKYLELNPTHLNALGWKS 426
EVRELVK AGLE++SLKRVRIGGF+LP DLG+GK++EL + L ALGWK+
Sbjct: 361 EVRELVKNAGLEVHSLKRVRIGGFRLPSDLGLGKHVELKQSELKALGWKN 410
>ref|NP_189920.1| putative protein; protein id: At3g43340.1 [Arabidopsis thaliana]
gi|11358365|pir||T47376 hypothetical protein T5C2.40 -
Arabidopsis thaliana gi|7263609|emb|CAB81575.1| putative
protein [Arabidopsis thaliana]
Length = 74
Score = 49.7 bits (117), Expect = 3e-05
Identities = 24/31 (77%), Positives = 26/31 (83%)
Frame = -3
Query: 575 EVRELVKRAGLEIYSLKRVRIGGFKLPPDLG 483
EVRELVK GLE+ SLKRVRIGG +LP DLG
Sbjct: 44 EVRELVKNVGLEVDSLKRVRIGGVRLPSDLG 74
>ref|NP_345185.1| ribosomal small subunit pseudouridine synthase A [Streptococcus
pneumoniae TIGR4] gi|25323992|pir||H95078 hypothetical
protein SP0680 [imported] - Streptococcus pneumoniae
(strain TIGR4) gi|14972155|gb|AAK74825.1| ribosomal
small subunit pseudouridine synthase A [Streptococcus
pneumoniae TIGR4]
Length = 241
Score = 37.7 bits (86), Expect = 0.10
Identities = 16/46 (34%), Positives = 27/46 (57%)
Frame = -3
Query: 575 EVRELVKRAGLEIYSLKRVRIGGFKLPPDLGIGKYLELNPTHLNAL 438
+++++ G+++ SLKR++ G F L PDL G Y LN L +
Sbjct: 188 QIKKMFLSVGVKVTSLKRIQFGDFTLNPDLAEGNYRPLNQKELQII 233
>gb|ZP_00035279.1| hypothetical protein [Enterococcus faecium]
Length = 239
Score = 37.4 bits (85), Expect = 0.14
Identities = 17/43 (39%), Positives = 26/43 (59%)
Frame = -3
Query: 575 EVRELVKRAGLEIYSLKRVRIGGFKLPPDLGIGKYLELNPTHL 447
+V+ +VK G E+ L+R+R+G F LP DL G+Y + L
Sbjct: 191 QVKRMVKAVGKEVLYLQRIRMGKFWLPKDLNKGQYRPMTKEEL 233
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,131,625
Number of Sequences: 1393205
Number of extensions: 10114654
Number of successful extensions: 20555
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 20144
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20547
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)