Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003952A_C01 KMC003952A_c01
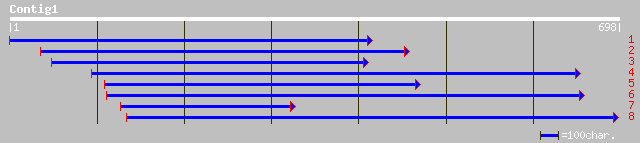
(691 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM61228.1| unknown [Arabidopsis thaliana] 163 3e-50
pir||A84779 hypothetical protein At2g36300 [imported] - Arabidop... 162 4e-50
ref|NP_565842.1| expressed protein; protein id: At2g36300.1, sup... 162 4e-50
ref|NP_190844.1| hypothetical protein; protein id: At3g52760.1 [... 156 9e-48
ref|NP_011688.1| Golgi integral membrane protein; binds to the t... 69 1e-12
>gb|AAM61228.1| unknown [Arabidopsis thaliana]
Length = 255
Score = 163 bits (412), Expect(2) = 3e-50
Identities = 81/104 (77%), Positives = 91/104 (86%), Gaps = 1/104 (0%)
Frame = -2
Query: 690 PILLYMAFCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRTGNLDLHTCTSVVGYC 511
PI LY+A CLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGR GNL+LHTCTS+VGYC
Sbjct: 124 PIFLYLALCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRNGNLNLHTCTSLVGYC 183
Query: 510 MFPVVIFSALSLFLPQG-GVFGLAIAAVFVLWSTRASTGLIVSL 382
+ PVVI SA+SLF+PQG G +AA+FVLWSTRA + L+VSL
Sbjct: 184 LLPVVILSAVSLFVPQGAGPVRFVLAALFVLWSTRACSTLVVSL 227
Score = 57.8 bits (138), Expect(2) = 3e-50
Identities = 26/27 (96%), Positives = 27/27 (99%)
Frame = -3
Query: 377 DGGDEHRGLIAYACFLIYTLFSLLVIF 297
DGG+EHRGLIAYACFLIYTLFSLLVIF
Sbjct: 229 DGGEEHRGLIAYACFLIYTLFSLLVIF 255
>pir||A84779 hypothetical protein At2g36300 [imported] - Arabidopsis thaliana
Length = 283
Score = 162 bits (411), Expect(2) = 4e-50
Identities = 80/104 (76%), Positives = 91/104 (86%), Gaps = 1/104 (0%)
Frame = -2
Query: 690 PILLYMAFCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRTGNLDLHTCTSVVGYC 511
PI LY+A CLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGR GNL+LHTCTS+VGYC
Sbjct: 152 PIFLYLALCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRNGNLNLHTCTSLVGYC 211
Query: 510 MFPVVIFSALSLFLPQG-GVFGLAIAAVFVLWSTRASTGLIVSL 382
+ PVV+ SA+SLF+PQG G +AA+FVLWSTRA + L+VSL
Sbjct: 212 LLPVVVLSAVSLFVPQGAGPVRFVLAALFVLWSTRACSTLVVSL 255
Score = 57.8 bits (138), Expect(2) = 4e-50
Identities = 26/27 (96%), Positives = 27/27 (99%)
Frame = -3
Query: 377 DGGDEHRGLIAYACFLIYTLFSLLVIF 297
DGG+EHRGLIAYACFLIYTLFSLLVIF
Sbjct: 257 DGGEEHRGLIAYACFLIYTLFSLLVIF 283
>ref|NP_565842.1| expressed protein; protein id: At2g36300.1, supported by cDNA:
114613. [Arabidopsis thaliana]
gi|20197939|gb|AAD21436.2| expressed protein
[Arabidopsis thaliana]
Length = 255
Score = 162 bits (411), Expect(2) = 4e-50
Identities = 80/104 (76%), Positives = 91/104 (86%), Gaps = 1/104 (0%)
Frame = -2
Query: 690 PILLYMAFCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRTGNLDLHTCTSVVGYC 511
PI LY+A CLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGR GNL+LHTCTS+VGYC
Sbjct: 124 PIFLYLALCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRNGNLNLHTCTSLVGYC 183
Query: 510 MFPVVIFSALSLFLPQG-GVFGLAIAAVFVLWSTRASTGLIVSL 382
+ PVV+ SA+SLF+PQG G +AA+FVLWSTRA + L+VSL
Sbjct: 184 LLPVVVLSAVSLFVPQGAGPVRFVLAALFVLWSTRACSTLVVSL 227
Score = 57.8 bits (138), Expect(2) = 4e-50
Identities = 26/27 (96%), Positives = 27/27 (99%)
Frame = -3
Query: 377 DGGDEHRGLIAYACFLIYTLFSLLVIF 297
DGG+EHRGLIAYACFLIYTLFSLLVIF
Sbjct: 229 DGGEEHRGLIAYACFLIYTLFSLLVIF 255
>ref|NP_190844.1| hypothetical protein; protein id: At3g52760.1 [Arabidopsis
thaliana] gi|11288410|pir||T49029 hypothetical protein
F3C22.160 - Arabidopsis thaliana
gi|7669950|emb|CAB89237.1| putative protein [Arabidopsis
thaliana]
Length = 257
Score = 156 bits (394), Expect(2) = 9e-48
Identities = 76/104 (73%), Positives = 88/104 (84%), Gaps = 1/104 (0%)
Frame = -2
Query: 690 PILLYMAFCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRTGNLDLHTCTSVVGYC 511
PI LY+A CLFQLLAGKIQFGVILGW+VVSSIFLY+VFNMLAGR GNL+LHTCTS+VGY
Sbjct: 126 PIFLYLALCLFQLLAGKIQFGVILGWVVVSSIFLYIVFNMLAGRNGNLNLHTCTSLVGYS 185
Query: 510 MFPVVIFSALSLFLPQG-GVFGLAIAAVFVLWSTRASTGLIVSL 382
+ PVVI SA+SLF+PQG G + A FVLW+TRA + L+VSL
Sbjct: 186 LLPVVILSAVSLFVPQGAGPVRFVLGAAFVLWATRACSNLVVSL 229
Score = 56.6 bits (135), Expect(2) = 9e-48
Identities = 25/27 (92%), Positives = 27/27 (99%)
Frame = -3
Query: 377 DGGDEHRGLIAYACFLIYTLFSLLVIF 297
DGG+EHRGLI+YACFLIYTLFSLLVIF
Sbjct: 231 DGGEEHRGLISYACFLIYTLFSLLVIF 257
>ref|NP_011688.1| Golgi integral membrane protein; binds to the transport GTPases
Ypt1p and Ypt31p; Yip1p [Saccharomyces cerevisiae]
gi|1724030|sp|P53039|YIPA_YEAST YIP1 PROTEIN
gi|2133212|pir||S64486 YIP1 protein - yeast
(Saccharomyces cerevisiae) gi|1279715|emb|CAA66031.1|
YIP1 [Saccharomyces cerevisiae]
gi|1323304|emb|CAA97198.1| ORF YGR172c [Saccharomyces
cerevisiae]
Length = 248
Score = 68.6 bits (166), Expect(2) = 1e-12
Identities = 34/107 (31%), Positives = 57/107 (52%), Gaps = 4/107 (3%)
Frame = -2
Query: 690 PILLYMAFCLFQLLAGKIQFGVILGWIVVSSIFLYVVFNMLAGRTG----NLDLHTCTSV 523
P++ ++ F LF L+AGK+ FG I G + +I L+ + +++ NL S+
Sbjct: 114 PLIFFLLFGLFLLMAGKVHFGYIYGVALFGTISLHNLSKLMSNNDTSTQTNLQFFNTASI 173
Query: 522 VGYCMFPVVIFSALSLFLPQGGVFGLAIAAVFVLWSTRASTGLIVSL 382
+GYC P+ S L +F G ++ +FV+WST S+G + SL
Sbjct: 174 LGYCFLPLCFLSLLGIFHGLNNTTGYVVSVLFVIWSTWTSSGFLNSL 220
Score = 26.2 bits (56), Expect(2) = 1e-12
Identities = 11/21 (52%), Positives = 16/21 (75%)
Frame = -3
Query: 359 RGLIAYACFLIYTLFSLLVIF 297
R LIAY + Y++F+L+VIF
Sbjct: 227 RLLIAYPLLIFYSVFALMVIF 247
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 632,560,741
Number of Sequences: 1393205
Number of extensions: 14710374
Number of successful extensions: 54241
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 48923
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 53719
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31118763720
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)