Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003947A_C01 KMC003947A_c01
(678 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
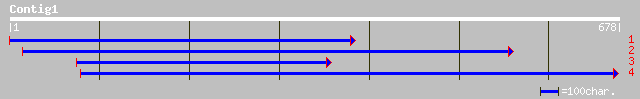
pir||G84711 hypothetical protein At2g30710 [imported] - Arabidop... 253 2e-66
ref|NP_565706.1| expressed protein; protein id: At2g30710.1, sup... 253 2e-66
emb|CAC18313.2| related to GTPase activating protein [Neurospora... 159 3e-38
gb|EAA31996.1| related to GTPase activating protein [MIPS] [Neur... 159 3e-38
ref|NP_595314.1| TBC domain protein. [Schizosaccharomyces pombe]... 155 4e-37
>pir||G84711 hypothetical protein At2g30710 [imported] - Arabidopsis thaliana
Length = 343
Score = 253 bits (646), Expect = 2e-66
Identities = 116/135 (85%), Positives = 129/135 (94%)
Frame = -1
Query: 678 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 499
QPGIQRLVFKLKELVRRID+PVS HME GLEFLQFAFRW+NCLLIREIPFN++ RLWDT
Sbjct: 209 QPGIQRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDT 268
Query: 498 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 319
YLAEGDALPDFLVYI+ASFLLTWSD+L+KLDFQ++VMFLQHLPT +W+ QELEMVLSRA+
Sbjct: 269 YLAEGDALPDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRAY 328
Query: 318 MWHSMFNNSPNHLAT 274
MWHSMFNNSPNHLA+
Sbjct: 329 MWHSMFNNSPNHLAS 343
>ref|NP_565706.1| expressed protein; protein id: At2g30710.1, supported by cDNA:
gi_13877620 [Arabidopsis thaliana]
gi|13877621|gb|AAK43888.1|AF370511_1 Unknown protein
[Arabidopsis thaliana] gi|20196901|gb|AAC02742.2|
expressed protein [Arabidopsis thaliana]
gi|22136274|gb|AAM91215.1| unknown protein [Arabidopsis
thaliana]
Length = 440
Score = 253 bits (646), Expect = 2e-66
Identities = 116/135 (85%), Positives = 129/135 (94%)
Frame = -1
Query: 678 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 499
QPGIQRLVFKLKELVRRID+PVS HME GLEFLQFAFRW+NCLLIREIPFN++ RLWDT
Sbjct: 306 QPGIQRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDT 365
Query: 498 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 319
YLAEGDALPDFLVYI+ASFLLTWSD+L+KLDFQ++VMFLQHLPT +W+ QELEMVLSRA+
Sbjct: 366 YLAEGDALPDFLVYIYASFLLTWSDELKKLDFQEMVMFLQHLPTHNWSDQELEMVLSRAY 425
Query: 318 MWHSMFNNSPNHLAT 274
MWHSMFNNSPNHLA+
Sbjct: 426 MWHSMFNNSPNHLAS 440
>emb|CAC18313.2| related to GTPase activating protein [Neurospora crassa]
Length = 602
Score = 159 bits (403), Expect = 3e-38
Identities = 68/133 (51%), Positives = 100/133 (75%)
Frame = -1
Query: 678 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 499
QPGIQR V L++L +RID ++ H+E + +EF+QF+FRW NCLL+REI R+WDT
Sbjct: 455 QPGIQRQVAALRDLTQRIDAGLAKHLEEENVEFIQFSFRWMNCLLMREISVKNTIRMWDT 514
Query: 498 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 319
Y+AE +F +Y+ A+FL+ WSDKL K+DFQ+++MFLQ LPT++WT +++E++LS A+
Sbjct: 515 YMAEEQGFSEFHLYVCAAFLVKWSDKLVKMDFQEIMMFLQSLPTREWTEKDIELLLSEAY 574
Query: 318 MWHSMFNNSPNHL 280
+W S+F S HL
Sbjct: 575 IWQSLFKGSSAHL 587
>gb|EAA31996.1| related to GTPase activating protein [MIPS] [Neurospora crassa]
Length = 600
Score = 159 bits (403), Expect = 3e-38
Identities = 68/133 (51%), Positives = 100/133 (75%)
Frame = -1
Query: 678 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 499
QPGIQR V L++L +RID ++ H+E + +EF+QF+FRW NCLL+REI R+WDT
Sbjct: 453 QPGIQRQVAALRDLTQRIDAGLAKHLEEENVEFIQFSFRWMNCLLMREISVKNTIRMWDT 512
Query: 498 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 319
Y+AE +F +Y+ A+FL+ WSDKL K+DFQ+++MFLQ LPT++WT +++E++LS A+
Sbjct: 513 YMAEEQGFSEFHLYVCAAFLVKWSDKLVKMDFQEIMMFLQSLPTREWTEKDIELLLSEAY 572
Query: 318 MWHSMFNNSPNHL 280
+W S+F S HL
Sbjct: 573 IWQSLFKGSSAHL 585
>ref|NP_595314.1| TBC domain protein. [Schizosaccharomyces pombe]
gi|7490611|pir||T40517 GTPase activating protein -
fission yeast (Schizosaccharomyces pombe)
gi|3150248|emb|CAA19167.1| TBC domain protein.
[Schizosaccharomyces pombe]
Length = 514
Score = 155 bits (393), Expect = 4e-37
Identities = 65/134 (48%), Positives = 105/134 (77%), Gaps = 1/134 (0%)
Frame = -1
Query: 678 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 499
QPGI+R V L+EL RID+P+ H++ +G++FLQF+FRW NCLL+RE+ + + R+WDT
Sbjct: 379 QPGIRRQVNNLRELTLRIDEPLVKHLQMEGVDFLQFSFRWMNCLLMRELSISNIIRMWDT 438
Query: 498 YLAEG-DALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRA 322
Y+AEG +F +Y+ A+FL+ WS +LQK++FQD+++FLQ +PT+DW+ +++E++LS A
Sbjct: 439 YMAEGVQGFSEFHLYVCAAFLVKWSSELQKMEFQDILIFLQSIPTKDWSTKDIEILLSEA 498
Query: 321 FMWHSMFNNSPNHL 280
F+W S+++ + HL
Sbjct: 499 FLWKSLYSGAGAHL 512
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,344,131
Number of Sequences: 1393205
Number of extensions: 13332372
Number of successful extensions: 32090
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 30944
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32033
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29987172312
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)