Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003938A_C01 KMC003938A_c01
(598 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
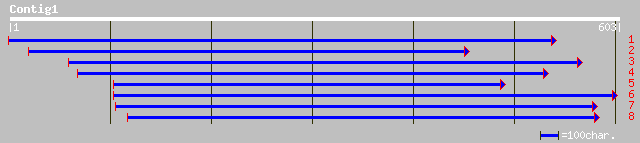
ref|NP_192632.1| putative protein disulfide isomerase; protein i... 62 7e-09
ref|NP_564452.1| expressed protein; protein id: At1g34780.1, sup... 52 6e-06
gb|AAO72589.1| disulfide isomerase-like protein [Oryza sativa (j... 51 1e-05
gb|EAA15463.1| C2 domain, putative [Plasmodium yoelii yoelii] 35 0.56
gb|EAA21900.1| hypothetical protein [Plasmodium yoelii yoelii] 34 1.2
>ref|NP_192632.1| putative protein disulfide isomerase; protein id: At4g08930.1
[Arabidopsis thaliana] gi|25372635|pir||A85090
hypothetical protein AT4g08930 [imported] - Arabidopsis
thaliana gi|4325372|gb|AAD17368.1| contains similarity
to protein disulfide isomerases [Arabidopsis thaliana]
gi|7267535|emb|CAB78017.1| putative protein disulfide
isomerase [Arabidopsis thaliana]
Length = 327
Score = 61.6 bits (148), Expect = 7e-09
Identities = 30/36 (83%), Positives = 31/36 (85%)
Frame = -1
Query: 595 LKEPCKRSNLQEGAFNAKAWASKSLATVSIGEESSS 488
LKEPC SNLQEGA NA+AWASKSLATVSI E SSS
Sbjct: 284 LKEPCMSSNLQEGAMNARAWASKSLATVSIAESSSS 319
>ref|NP_564452.1| expressed protein; protein id: At1g34780.1, supported by cDNA:
gi_16649012, supported by cDNA: gi_20260019 [Arabidopsis
thaliana] gi|5668777|gb|AAD46003.1|AC007894_1 Contains
similarity to gi|729436 protein disulfide
isomerase-related protein precursor from Rattus
norvegicus. [Arabidopsis thaliana]
gi|16649013|gb|AAL24358.1| Unknown protein [Arabidopsis
thaliana] gi|20260020|gb|AAM13357.1| unknown protein
[Arabidopsis thaliana]
Length = 310
Score = 52.0 bits (123), Expect = 6e-06
Identities = 25/31 (80%), Positives = 28/31 (89%)
Frame = -1
Query: 580 KRSNLQEGAFNAKAWASKSLATVSIGEESSS 488
+RSNLQ GA NA+AWASKSLATVSIG+ SSS
Sbjct: 272 RRSNLQGGAMNARAWASKSLATVSIGDSSSS 302
>gb|AAO72589.1| disulfide isomerase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 311
Score = 50.8 bits (120), Expect = 1e-05
Identities = 25/32 (78%), Positives = 27/32 (84%)
Frame = -1
Query: 586 PCKRSNLQEGAFNAKAWASKSLATVSIGEESS 491
P KR NLQEGA NA AWASKSLA+VSIGE S+
Sbjct: 269 PSKRGNLQEGARNATAWASKSLASVSIGEPST 300
>gb|EAA15463.1| C2 domain, putative [Plasmodium yoelii yoelii]
Length = 717
Score = 35.4 bits (80), Expect = 0.56
Identities = 30/118 (25%), Positives = 51/118 (42%), Gaps = 5/118 (4%)
Frame = +2
Query: 41 SHTYX--VGNFTNNHT*TWYPISTRQELQRFHSQQK*IYDDQKI*PSKKTYKSKN---YR 205
+HTY + N TN + IS + + Q +++ +K + + I KKTYK N R
Sbjct: 179 THTYKKDISNNTNINKENEKVISKKNDEQNYNTLEKNSFSNNNIKSLKKTYKEPNEYSVR 238
Query: 206 LHLKFAKFKLYTLKSQFSRKLNEVFRDHIDKSRSITSFSNIIDLISMRFKGVKMMLDT 379
+ K Y + S ++ + IDK+R I ++ + K +K DT
Sbjct: 239 IIAKLINKTKYNVFSYILNSMSSLNYRCIDKTRGILDAELNVNKLYEELKVIKFNADT 296
>gb|EAA21900.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1918
Score = 34.3 bits (77), Expect = 1.2
Identities = 19/46 (41%), Positives = 24/46 (51%)
Frame = +2
Query: 62 NFTNNHT*TWYPISTRQELQRFHSQQK*IYDDQKI*PSKKTYKSKN 199
N T+NHT + I T Q+LQ HS Y D+K K K+KN
Sbjct: 899 NDTDNHTHKYSKIRTDQKLQNMHSNNTSNYSDEKKKNKDKCQKNKN 944
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 463,100,256
Number of Sequences: 1393205
Number of extensions: 9079368
Number of successful extensions: 19315
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 18884
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19314
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23140425222
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)