Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003930A_C01 KMC003930A_c01
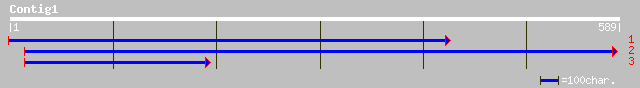
(588 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T01333 hypothetical protein F6N15.11 - Arabidopsis thaliana... 117 8e-26
ref|NP_191916.2| putative transcriptional regulator; protein id:... 117 8e-26
dbj|BAC41905.1| putative bHLH transcription factor bHLH016 [Arab... 117 8e-26
gb|AAM10933.1|AF488561_1 putative bHLH transcription factor [Ara... 116 2e-25
gb|AAK55467.1|AC069300_22 putative DNA-binding protein [Oryza sa... 66 3e-10
>pir||T01333 hypothetical protein F6N15.11 - Arabidopsis thaliana
gi|3193326|gb|AAC19308.1| contains similarity to
transcriptional activators such as Ra-like and myc-like
regulatory R proteins [Arabidopsis thaliana]
gi|7267092|emb|CAB80763.1| putative transcriptional
regulator [Arabidopsis thaliana]
Length = 329
Score = 117 bits (294), Expect = 8e-26
Identities = 71/135 (52%), Positives = 91/135 (66%), Gaps = 9/135 (6%)
Frame = -2
Query: 587 NMSSMMLPMTMQQQ--LQMSIMG-PMGMGMGMGMGMGMGMGMDMNTMNRA--NMPGMPPV 423
NM SMMLPM MQQQ LQMS+M PMG+GMGMGM G+G+ +D+N+MNRA + P +
Sbjct: 203 NMPSMMLPMAMQQQQQLQMSLMSNPMGLGMGMGMP-GLGL-LDLNSMNRAAASAPNIHAN 260
Query: 422 LHPSAFMPM--ASWDAATAATGGDRLQGPPATMMPDPMSTFFGCQSQPMNMEAYSRLAAM 249
+ P+ F+PM SWDA++ R Q P ++PDPMS F C +QP MEAYSR+A +
Sbjct: 261 MMPNPFLPMNCPSWDASS---NDSRFQSP---LIPDPMSAFLACSTQPTTMEAYSRMATL 314
Query: 248 YQQLHQ--PPASGPK 210
YQQ+ Q PP S PK
Sbjct: 315 YQQMQQQLPPPSNPK 329
>ref|NP_191916.2| putative transcriptional regulator; protein id: At4g00050.1
[Arabidopsis thaliana]
Length = 395
Score = 117 bits (294), Expect = 8e-26
Identities = 71/135 (52%), Positives = 91/135 (66%), Gaps = 9/135 (6%)
Frame = -2
Query: 587 NMSSMMLPMTMQQQ--LQMSIMG-PMGMGMGMGMGMGMGMGMDMNTMNRA--NMPGMPPV 423
NM SMMLPM MQQQ LQMS+M PMG+GMGMGM G+G+ +D+N+MNRA + P +
Sbjct: 269 NMPSMMLPMAMQQQQQLQMSLMSNPMGLGMGMGMP-GLGL-LDLNSMNRAAASAPNIHAN 326
Query: 422 LHPSAFMPM--ASWDAATAATGGDRLQGPPATMMPDPMSTFFGCQSQPMNMEAYSRLAAM 249
+ P+ F+PM SWDA++ R Q P ++PDPMS F C +QP MEAYSR+A +
Sbjct: 327 MMPNPFLPMNCPSWDASS---NDSRFQSP---LIPDPMSAFLACSTQPTTMEAYSRMATL 380
Query: 248 YQQLHQ--PPASGPK 210
YQQ+ Q PP S PK
Sbjct: 381 YQQMQQQLPPPSNPK 395
>dbj|BAC41905.1| putative bHLH transcription factor bHLH016 [Arabidopsis thaliana]
Length = 399
Score = 117 bits (294), Expect = 8e-26
Identities = 71/135 (52%), Positives = 91/135 (66%), Gaps = 9/135 (6%)
Frame = -2
Query: 587 NMSSMMLPMTMQQQ--LQMSIMG-PMGMGMGMGMGMGMGMGMDMNTMNRA--NMPGMPPV 423
NM SMMLPM MQQQ LQMS+M PMG+GMGMGM G+G+ +D+N+MNRA + P +
Sbjct: 273 NMPSMMLPMAMQQQQQLQMSLMSNPMGLGMGMGMP-GLGL-LDLNSMNRAAASAPNIHAN 330
Query: 422 LHPSAFMPM--ASWDAATAATGGDRLQGPPATMMPDPMSTFFGCQSQPMNMEAYSRLAAM 249
+ P+ F+PM SWDA++ R Q P ++PDPMS F C +QP MEAYSR+A +
Sbjct: 331 MMPNPFLPMNCPSWDASS---NDSRFQSP---LIPDPMSAFLACSTQPTTMEAYSRMATL 384
Query: 248 YQQLHQ--PPASGPK 210
YQQ+ Q PP S PK
Sbjct: 385 YQQMQQQLPPPSNPK 399
>gb|AAM10933.1|AF488561_1 putative bHLH transcription factor [Arabidopsis thaliana]
Length = 399
Score = 116 bits (290), Expect = 2e-25
Identities = 70/135 (51%), Positives = 91/135 (66%), Gaps = 9/135 (6%)
Frame = -2
Query: 587 NMSSMMLPMTMQQQ--LQMSIMG-PMGMGMGMGMGMGMGMGMDMNTMNRA--NMPGMPPV 423
NM SMMLPM MQQQ LQMS+M PMG+GMGMGM G+G+ +D+N+MNRA + P +
Sbjct: 273 NMPSMMLPMAMQQQQQLQMSLMSNPMGLGMGMGMP-GLGL-LDLNSMNRAAASAPNIHAN 330
Query: 422 LHPSAFMPM--ASWDAATAATGGDRLQGPPATMMPDPMSTFFGCQSQPMNMEAYSRLAAM 249
+ P+ F+PM SWDA++ R Q P ++PDPMS F C +QP MEAYSR+A +
Sbjct: 331 MMPNPFLPMNCPSWDASS---NDSRFQSP---LIPDPMSAFLACSTQPTTMEAYSRMATL 384
Query: 248 YQQLHQ--PPASGPK 210
YQQ+ + PP S PK
Sbjct: 385 YQQMQRQLPPPSNPK 399
>gb|AAK55467.1|AC069300_22 putative DNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 191
Score = 66.2 bits (160), Expect = 3e-10
Identities = 48/128 (37%), Positives = 63/128 (48%), Gaps = 11/128 (8%)
Frame = -2
Query: 584 MSSMMLPMTMQQ-QLQMSIMGPMGMGMGMGMGMGMGMGMDMNTMNRANMPGMPPV-LHPS 411
M SMM+PM M QLQMS+M M +G+ M MN G P+ +H
Sbjct: 60 MGSMMMPMGMAMPQLQMSVMAQMAQMAQIGLSM----------MNMGQAGGYAPMHMHTP 109
Query: 410 AFMPMASWDAA---TAATGGDRLQGPPATMMPDPMSTFFGCQS------QPMNMEAYSRL 258
F+P+ SWDAA ++A DR P D S F Q+ QP MEAY+R+
Sbjct: 110 PFLPV-SWDAAASSSSAAAADRPPQPTGAATSDAFSAFLASQAAQQNAQQPNGMEAYNRM 168
Query: 257 AAMYQQLH 234
AMYQ+L+
Sbjct: 169 MAMYQKLN 176
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 623,937,443
Number of Sequences: 1393205
Number of extensions: 16922230
Number of successful extensions: 167397
Number of sequences better than 10.0: 1288
Number of HSP's better than 10.0 without gapping: 64947
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 125843
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22283372436
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)