Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003923A_C01 KMC003923A_c01
(642 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
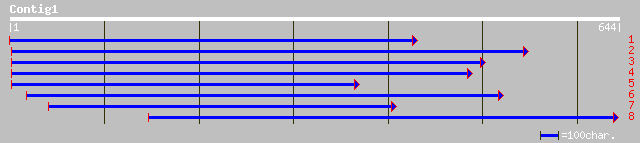
gb|AAM98205.1| putative protein [Arabidopsis thaliana] 105 5e-22
ref|NP_182050.1| unknown protein; protein id: At2g45250.1 [Arabi... 98 8e-20
ref|NP_195541.1| putative protein; protein id: At4g38280.1 [Arab... 80 2e-14
ref|NP_640426.1| ankyrin-like protein [Xanthomonas axonopodis pv... 35 1.1
gb|AAO51092.1| similar to Dictyostelium discoideum (Slime mold).... 35 1.1
>gb|AAM98205.1| putative protein [Arabidopsis thaliana]
Length = 170
Score = 105 bits (262), Expect = 5e-22
Identities = 51/80 (63%), Positives = 66/80 (81%)
Frame = -1
Query: 528 MQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQ 349
+ WEERY LQMLL KL+QSD+ ++VQML SLSS ELS+HAV+LEKR+IQ SLEEA+E+Q
Sbjct: 90 LDWEERYLHLQMLLNKLNQSDRTDHVQMLWSLSSAELSKHAVDLEKRSIQFSLEEAREMQ 149
Query: 348 RVAVLNVLGKSAKKFKAPAD 289
RVA LN+LG+S K+ ++
Sbjct: 150 RVAALNMLGRSVNSLKSTSN 169
>ref|NP_182050.1| unknown protein; protein id: At2g45250.1 [Arabidopsis thaliana]
gi|25341933|pir||C84888 hypothetical protein At2g45250
[imported] - Arabidopsis thaliana
gi|2583136|gb|AAB82645.1| unknown protein [Arabidopsis
thaliana]
Length = 211
Score = 98.2 bits (243), Expect = 8e-20
Identities = 51/87 (58%), Positives = 66/87 (75%), Gaps = 7/87 (8%)
Frame = -1
Query: 528 MQWEERYQQLQMLLRKLDQSDQVEYVQ-------MLQSLSSVELSRHAVELEKRAIQLSL 370
+ WEERY LQMLL KL+QSD+ ++VQ +L SLSS ELS+HAV+LEKR+IQ SL
Sbjct: 124 LDWEERYLHLQMLLNKLNQSDRTDHVQNMFPLLLVLWSLSSAELSKHAVDLEKRSIQFSL 183
Query: 369 EEAKELQRVAVLNVLGKSAKKFKAPAD 289
EEA+E+QRVA LNVLG+S K+ ++
Sbjct: 184 EEAREMQRVAALNVLGRSVNSIKSTSN 210
>ref|NP_195541.1| putative protein; protein id: At4g38280.1 [Arabidopsis thaliana]
gi|7485959|pir||T05656 hypothetical protein F22I13.50 -
Arabidopsis thaliana gi|4539336|emb|CAB37484.1| putative
protein [Arabidopsis thaliana]
gi|7270812|emb|CAB80493.1| putative protein [Arabidopsis
thaliana]
Length = 159
Score = 80.5 bits (197), Expect = 2e-14
Identities = 39/55 (70%), Positives = 47/55 (84%)
Frame = -1
Query: 528 MQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEE 364
+ WEERY LQMLL KL+QSD+ ++VQML SLSS ELS+HAV+LEKR+IQ SLEE
Sbjct: 90 LDWEERYLHLQMLLNKLNQSDRTDHVQMLWSLSSAELSKHAVDLEKRSIQFSLEE 144
>ref|NP_640426.1| ankyrin-like protein [Xanthomonas axonopodis pv. citri str. 306]
gi|21106115|gb|AAM34962.1| ankyrin-like protein
[Xanthomonas axonopodis pv. citri str. 306]
Length = 573
Score = 34.7 bits (78), Expect = 1.1
Identities = 30/94 (31%), Positives = 42/94 (43%)
Frame = -1
Query: 570 SSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEK 391
S GP + K Q E QQ++ + + Q Q +Y + LSRH VEL K
Sbjct: 474 SLGPELAQAKQEAQQQAREAQQQIRDVAQ---QHRQAQYAYAAAVRQADALSRHQVELAK 530
Query: 390 RAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPAD 289
+A EA+ QR A + A+K +A AD
Sbjct: 531 QAALQGRAEARRGQREAAQAQV--EAEKAQAEAD 562
>gb|AAO51092.1| similar to Dictyostelium discoideum (Slime mold). Interaptin
Length = 1781
Score = 34.7 bits (78), Expect = 1.1
Identities = 24/61 (39%), Positives = 37/61 (60%), Gaps = 2/61 (3%)
Frame = -1
Query: 525 QWEERYQQLQMLLRKL-DQS-DQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKEL 352
Q++E Q L + L KL DQS DQ+E +Q LQS +S++L ++ +Q SL E +L
Sbjct: 919 QYQETCQSLSLKLSKLNDQSNDQLEQIQQLQSSNSLDLQNSQNQIS--LLQDSLNETSDL 976
Query: 351 Q 349
+
Sbjct: 977 K 977
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,560,088
Number of Sequences: 1393205
Number of extensions: 9924973
Number of successful extensions: 29745
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 28595
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29714
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27007650415
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)