Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003916A_C01 KMC003916A_c01
(573 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
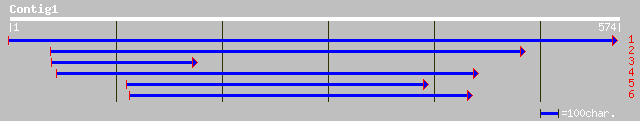
ref|NP_198138.1| putative protein; protein id: At5g27860.1 [Arab... 44 0.002
ref|NP_788624.1| CG33208-PF [Drosophila melanogaster] gi|2857311... 34 1.1
dbj|BAC26862.1| unnamed protein product [Mus musculus] 32 5.7
sp|P19593|RDPO_SCEOB Probable reverse transcriptase gi|81302|pir... 31 9.7
>ref|NP_198138.1| putative protein; protein id: At5g27860.1 [Arabidopsis thaliana]
Length = 224
Score = 43.5 bits (101), Expect = 0.002
Identities = 29/58 (50%), Positives = 35/58 (60%)
Frame = -1
Query: 567 KEKQHDERSSSPVQLSKFLARGKDDGVKDDGVRRSAVSGKKILLKLEKTKEDKAAESQ 394
+ K+H R KFL R KDDG RRSAVSGKK+ +K+KEDKAAES+
Sbjct: 103 RHKRHKNRER------KFLNRDKDDGE-----RRSAVSGKKV----DKSKEDKAAESK 145
>ref|NP_788624.1| CG33208-PF [Drosophila melanogaster] gi|28573115|ref|NP_788622.1|
CG33208-PC [Drosophila melanogaster]
gi|28573117|ref|NP_788626.1| CG33208-PH [Drosophila
melanogaster] gi|28573119|ref|NP_788625.1| CG33208-PG
[Drosophila melanogaster] gi|28573121|ref|NP_788623.1|
CG33208-PD [Drosophila melanogaster]
gi|28381223|gb|AAO41533.1| CG33208-PC [Drosophila
melanogaster] gi|28381224|gb|AAO41534.1| CG33208-PD
[Drosophila melanogaster] gi|28381225|gb|AAO41535.1|
CG33208-PF [Drosophila melanogaster]
gi|28381226|gb|AAO41536.1| CG33208-PG [Drosophila
melanogaster] gi|28381227|gb|AAO41537.1| CG33208-PH
[Drosophila melanogaster]
Length = 4723
Score = 34.3 bits (77), Expect = 1.1
Identities = 21/48 (43%), Positives = 28/48 (57%)
Frame = +2
Query: 401 SAALSSFVFSSFRRIFLPDTALRRTPSSLTPSSLPRAKNLESCTGLLL 544
++A SS+ S+ R L D RR+P+S T S+L N ESC GL L
Sbjct: 2980 ASATSSYYPSTTRSSHLSDLFRRRSPASGTVSALSGYGNKESCIGLAL 3027
>dbj|BAC26862.1| unnamed protein product [Mus musculus]
Length = 389
Score = 32.0 bits (71), Expect = 5.7
Identities = 17/55 (30%), Positives = 26/55 (46%), Gaps = 9/55 (16%)
Frame = +3
Query: 111 WWPKTGNPTK----AQHIRGIHGSRTTFQQ-----TQAQSFHCKRNLNVSPSPFS 248
WWP+T P + Q +R I +TF + FHC R L++ P P++
Sbjct: 20 WWPQTSRPRRIQAVLQRLRAICPPLSTFYLFFVIFVVSTIFHCHRRLSLVPGPWA 74
>sp|P19593|RDPO_SCEOB Probable reverse transcriptase gi|81302|pir||S05341 probable
reverse transcriptase - green alga KS3/2 chloroplast
Length = 608
Score = 31.2 bits (69), Expect = 9.7
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 7/70 (10%)
Frame = +3
Query: 210 CKRNLNVSPSPFSL--TLPTKC*SACIL*GKKI*APNK*T*LCCTPKHTLIKTSI*K--- 374
C++NL ++ P+ L LP + GK PN LC +P H L+ +SI K
Sbjct: 528 CRKNLEINSIPYELHHILPKR------FGGKD--TPNNMVLLCKSPCHQLVSSSIQKADV 579
Query: 375 --VQQFISLG 398
+Q +ISLG
Sbjct: 580 SEIQNYISLG 589
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 493,212,742
Number of Sequences: 1393205
Number of extensions: 10488436
Number of successful extensions: 27384
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 26430
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27361
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)