Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC003884A_C01 KMC003884A_c01
(605 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
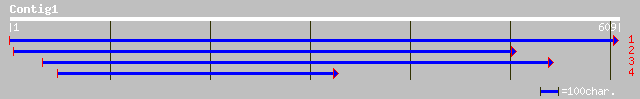
ref|NP_197191.1| Exostosin family; protein id: At5g16890.1 [Arab... 182 7e-47
dbj|BAB85236.1| P0425G02.11 [Oryza sativa (japonica cultivar-gro... 129 3e-31
ref|NP_181055.2| Exostosin family; protein id: At2g35100.1 [Arab... 91 9e-18
dbj|BAC55656.1| OJ1092_A07.9 [Oryza sativa (japonica cultivar-gr... 85 4e-16
ref|NP_199306.1| Exostosin family; protein id: At5g44930.1, supp... 84 2e-15
>ref|NP_197191.1| Exostosin family; protein id: At5g16890.1 [Arabidopsis thaliana]
gi|11357737|pir||T51544 hypothetical protein F2K13_40 -
Arabidopsis thaliana gi|9755690|emb|CAC01702.1| putative
protein [Arabidopsis thaliana]
gi|15810401|gb|AAL07088.1| unknown protein [Arabidopsis
thaliana] gi|23296585|gb|AAN13125.1| unknown protein
[Arabidopsis thaliana]
Length = 511
Score = 182 bits (461), Expect(2) = 7e-47
Identities = 82/120 (68%), Positives = 103/120 (85%)
Frame = -3
Query: 567 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 388
CIP IVSDELE PFEGILDY+K+A+ VSS DA++PGWL+ L+ + P Q+K LQ NLAQY
Sbjct: 384 CIPVIVSDELEFPFEGILDYKKVAVLVSSSDAIQPGWLVNHLRSLTPFQVKGLQNNLAQY 443
Query: 387 SRHFVFSNPAQPLGPEDLVWKMMAGKVVNIKLHSRRSQRVVEGSRNLCTCECRTGNITSS 208
SRHF++S+PAQPLGPEDL W+M+AGK+VNIKLH+RRSQRVV+GSR++C C+C N T+S
Sbjct: 444 SRHFLYSSPAQPLGPEDLTWRMIAGKLVNIKLHTRRSQRVVKGSRSICRCDCWRSNSTAS 503
Score = 27.3 bits (59), Expect(2) = 7e-47
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = -1
Query: 605 PSSARLYDAIVSG 567
PSSARL+DAIVSG
Sbjct: 371 PSSARLFDAIVSG 383
>dbj|BAB85236.1| P0425G02.11 [Oryza sativa (japonica cultivar-group)]
gi|20160709|dbj|BAB89652.1| P0468B07.31 [Oryza sativa
(japonica cultivar-group)]
Length = 480
Score = 129 bits (325), Expect(2) = 3e-31
Identities = 57/82 (69%), Positives = 73/82 (88%)
Frame = -3
Query: 567 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 388
CIP IVSDELELPFEGILDYRKIALFVSS DA++PGWL+K+L+ + +I+++Q NL +Y
Sbjct: 379 CIPVIVSDELELPFEGILDYRKIALFVSSNDAVQPGWLVKYLRSIDAKRIRQMQSNLLKY 438
Query: 387 SRHFVFSNPAQPLGPEDLVWKM 322
SRHF++S+PA+PLGPEDL W+M
Sbjct: 439 SRHFLYSSPARPLGPEDLTWRM 460
Score = 27.3 bits (59), Expect(2) = 3e-31
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = -1
Query: 605 PSSARLYDAIVSG 567
PSSARL+DAIVSG
Sbjct: 366 PSSARLFDAIVSG 378
>ref|NP_181055.2| Exostosin family; protein id: At2g35100.1 [Arabidopsis thaliana]
Length = 447
Score = 90.5 bits (223), Expect(2) = 9e-18
Identities = 51/125 (40%), Positives = 78/125 (61%), Gaps = 6/125 (4%)
Frame = -3
Query: 567 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 388
C+P IVSD +ELPFE ++DYRK ++FV + AL+PG+L++ L+ ++ +I E Q +
Sbjct: 327 CVPLIVSDSIELPFEDVIDYRKFSIFVEANAALQPGFLVQMLRKIKTKKILEYQREMKSV 386
Query: 387 SRHFVFSNPAQPLGPEDLVWKMMAGKVVNIKLHSRRSQRVVEGSRNL----CTCEC--RT 226
R+F + NP G +W+ ++ K+ IKL S R +R+V RNL C+C C +T
Sbjct: 387 RRYFDYDNPN---GAVKEIWRQVSHKLPLIKLMSNRDRRLV--LRNLTEPNCSCLCTNQT 441
Query: 225 GNITS 211
G ITS
Sbjct: 442 GLITS 446
Score = 21.2 bits (43), Expect(2) = 9e-18
Identities = 8/12 (66%), Positives = 11/12 (91%)
Frame = -1
Query: 605 PSSARLYDAIVS 570
PS+ RL+D+IVS
Sbjct: 314 PSACRLFDSIVS 325
>dbj|BAC55656.1| OJ1092_A07.9 [Oryza sativa (japonica cultivar-group)]
Length = 453
Score = 84.7 bits (208), Expect(2) = 4e-16
Identities = 41/112 (36%), Positives = 65/112 (57%)
Frame = -3
Query: 567 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 388
C+P IVSD +ELPFE ++DYR I++FV + A++PG+L L+ + +I E Q + +
Sbjct: 334 CVPVIVSDYIELPFEDVIDYRNISIFVETSKAVQPGFLTSTLRGISSQRILEYQREIKKV 393
Query: 387 SRHFVFSNPAQPLGPEDLVWKMMAGKVVNIKLHSRRSQRVVEGSRNLCTCEC 232
+F + +P GP + +W ++ K IKL R +R+VE N C C
Sbjct: 394 KHYFEYEDPN---GPVNQIWHQVSSKAPLIKLLINRDKRLVERGTNGTDCSC 442
Score = 21.6 bits (44), Expect(2) = 4e-16
Identities = 8/12 (66%), Positives = 11/12 (91%)
Frame = -1
Query: 605 PSSARLYDAIVS 570
PS+ RL+DA+VS
Sbjct: 321 PSACRLFDALVS 332
>ref|NP_199306.1| Exostosin family; protein id: At5g44930.1, supported by cDNA:
gi_15081732 [Arabidopsis thaliana]
gi|10177484|dbj|BAB10875.1|
gb|AAC61825.1~gene_id:K21C13.11~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|15081733|gb|AAK82521.1| AT5g44930/K21C13_11
[Arabidopsis thaliana] gi|27363262|gb|AAO11550.1|
At5g44930/K21C13_11 [Arabidopsis thaliana]
Length = 443
Score = 83.6 bits (205), Expect = 2e-15
Identities = 42/114 (36%), Positives = 72/114 (62%), Gaps = 2/114 (1%)
Frame = -3
Query: 567 CIPAIVSDELELPFEGILDYRKIALFVSSIDALKPGWLLKFLKDVRPAQIKELQPNLAQY 388
C+P IVSD +ELPFE ++DYRK ++F+ ALKPG+++K L+ V+P +I + Q + +
Sbjct: 323 CVPVIVSDGIELPFEDVIDYRKFSIFLRRDAALKPGFVVKKLRKVKPGKILKYQKVMKEV 382
Query: 387 SRHFVFSNPAQPLGPEDLVWKMMAGKVVNIKLHSRRSQRVV--EGSRNLCTCEC 232
R+F +++ G + +W+ + K+ IKL R +R++ +GS C+C C
Sbjct: 383 RRYFDYTHLN---GSVNEIWRQVTKKIPLIKLMINREKRMIKRDGSDPQCSCLC 433
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,437,634
Number of Sequences: 1393205
Number of extensions: 10493662
Number of successful extensions: 28811
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 27977
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 28789
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)