Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
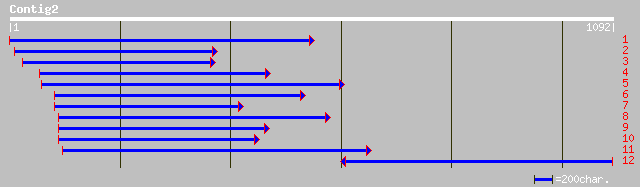
Query= KMC003824A_C02 KMC003824A_c02
(1077 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA09228.1| thaumatin-like protein PR-5b [Cicer arietinum] 454 e-126
gb|AAK59275.1|AF378571_1 thaumatin-like protein [Sambucus nigra] 369 e-101
gb|AAO13658.1| osmotin-like protein linusitin [Linum usitatissimum] 362 4e-99
ref|NP_192902.1| osmotin precursor; protein id: At4g11650.1, sup... 362 7e-99
gb|AAM61750.1| osmotin precursor [Arabidopsis thaliana] 362 7e-99
>emb|CAA09228.1| thaumatin-like protein PR-5b [Cicer arietinum]
Length = 239
Score = 454 bits (1168), Expect = e-126
Identities = 202/230 (87%), Positives = 215/230 (92%)
Frame = -1
Query: 1053 LTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVNAGTA 874
+TL SLL +LT S +AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNL VNAGT+
Sbjct: 7 ITLCSLLFLLTPS-----QAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLWVNAGTS 61
Query: 873 MARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLV 694
MARIWGRTGCNFDG+GRGRC+TGDCTGGL C GWGVPPNTLAEFALNQYGN DFYDISLV
Sbjct: 62 MARIWGRTGCNFDGSGRGRCETGDCTGGLQCTGWGVPPNTLAEFALNQYGNLDFYDISLV 121
Query: 693 DGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQGTC 514
DGFNIPMDFFP+NGGCHKISCTADINGQCPNELRA GGCNNPCTVYKTNE+CCTNGQG+C
Sbjct: 122 DGFNIPMDFFPINGGCHKISCTADINGQCPNELRAQGGCNNPCTVYKTNEYCCTNGQGSC 181
Query: 513 GPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPHVTL 364
GPTNFS FFKDRCHD+YSYPQDDPTSTFTCPAGSNYKVVFCPLG+PH+ +
Sbjct: 182 GPTNFSTFFKDRCHDAYSYPQDDPTSTFTCPAGSNYKVVFCPLGAPHIEM 231
>gb|AAK59275.1|AF378571_1 thaumatin-like protein [Sambucus nigra]
Length = 226
Score = 369 bits (947), Expect = e-101
Identities = 164/226 (72%), Positives = 187/226 (82%), Gaps = 1/226 (0%)
Frame = -1
Query: 1062 MGYLTLSSLLLMLTASLVTLTR-AANFEIVNNCPYTVWAAASPGGGRRLDRGQTWNLGVN 886
MG+L + + +L++ + AANF I NNCP+TVWAAA PGGGR+L+RGQ W+L V
Sbjct: 1 MGFLKSLPISIFFVIALISSSAYAANFNIRNNCPFTVWAAAVPGGGRQLNRGQVWSLDVP 60
Query: 885 AGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYD 706
AGT ARIW RT CNFDGAGRGRCQTGDC G L+CQ +G PPNTLAE+ALNQ+ N DF+D
Sbjct: 61 AGTRGARIWARTNCNFDGAGRGRCQTGDCNGLLSCQAYGAPPNTLAEYALNQFNNLDFFD 120
Query: 705 ISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNG 526
ISLVDGFN+ MDF P G I CTADINGQCPNELRAPGGCNNPCTVY+TNE+CCTNG
Sbjct: 121 ISLVDGFNVAMDFSPTGGCARGIQCTADINGQCPNELRAPGGCNNPCTVYRTNEYCCTNG 180
Query: 525 QGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 388
QGTCGPTNFSRFFK+RC D+YSYPQDDPTSTFTCP G+NY+VVFCP
Sbjct: 181 QGTCGPTNFSRFFKERCRDAYSYPQDDPTSTFTCPGGTNYRVVFCP 226
>gb|AAO13658.1| osmotin-like protein linusitin [Linum usitatissimum]
Length = 263
Score = 362 bits (930), Expect = 4e-99
Identities = 166/230 (72%), Positives = 188/230 (81%), Gaps = 8/230 (3%)
Frame = -1
Query: 1038 LLLMLTASL---VTLTRAANFEIVNNCPYTVWAAASP-GGGRRLDRGQTWNLGVNAGTAM 871
LL+ L +SL V++T AA +I NNCPYTVWAA++P GGGR+LD GQTW + AGT+M
Sbjct: 7 LLIPLFSSLLLWVSITNAAVIDIFNNCPYTVWAASTPIGGGRQLDHGQTWTIYPPAGTSM 66
Query: 870 ARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQDFYDISLVD 691
ARIWGR CNFDG+GRG C+TGDC G LNCQGWGVPPNTLAE+ALNQ+ N DFYDISLVD
Sbjct: 67 ARIWGRRNCNFDGSGRGWCETGDCGGVLNCQGWGVPPNTLAEYALNQFSNLDFYDISLVD 126
Query: 690 GFNIPMDFFPL----NGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFCCTNGQ 523
GFNIPM F P +G C ++CTADIN QCP ELRAPGGCNNPCTV+KTNE+CCT G
Sbjct: 127 GFNIPMIFTPTANVGSGNCQSLTCTADINTQCPGELRAPGGCNNPCTVFKTNEYCCTQGY 186
Query: 522 GTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPLGSPH 373
GTCGPT FSRFFKDRC SYSYPQDDP+STFTCP G+NY+VVFCP GS H
Sbjct: 187 GTCGPTGFSRFFKDRCPTSYSYPQDDPSSTFTCPGGTNYRVVFCPYGSTH 236
>ref|NP_192902.1| osmotin precursor; protein id: At4g11650.1, supported by cDNA: 13796.
[Arabidopsis thaliana] gi|21542444|sp|P50700|OSL3_ARATH
Osmotin-like protein OSM34 precursor
gi|7442160|pir||T04212 osmotin precursor - Arabidopsis
thaliana gi|4539456|emb|CAB39936.1| osmotin precursor
[Arabidopsis thaliana] gi|7267865|emb|CAB78208.1| osmotin
precursor [Arabidopsis thaliana]
Length = 244
Score = 362 bits (928), Expect = 7e-99
Identities = 163/230 (70%), Positives = 187/230 (80%)
Frame = -1
Query: 1077 LKSLEMGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWN 898
+ +L + S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W
Sbjct: 1 MANLLVSTFIFSALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWR 54
Query: 897 LGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQ 718
L V AGT MARIWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N
Sbjct: 55 LDVAAGTKMARIWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGQPPNTLAEYALNQFNNL 114
Query: 717 DFYDISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFC 538
DFYDISLVDGFNIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++C
Sbjct: 115 DFYDISLVDGFNIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYC 174
Query: 537 CTNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 388
CTNGQG+C T +SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 175 CTNGQGSCSDTEYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
>gb|AAM61750.1| osmotin precursor [Arabidopsis thaliana]
Length = 244
Score = 362 bits (928), Expect = 7e-99
Identities = 163/230 (70%), Positives = 188/230 (80%)
Frame = -1
Query: 1077 LKSLEMGYLTLSSLLLMLTASLVTLTRAANFEIVNNCPYTVWAAASPGGGRRLDRGQTWN 898
+ +L + S+LLL+ TA+ AA FEI+N C YTVWAAASPGGGRRLD GQ+W
Sbjct: 1 MANLLVSTFIFSALLLISTAT------AATFEILNQCSYTVWAAASPGGGRRLDAGQSWR 54
Query: 897 LGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDCTGGLNCQGWGVPPNTLAEFALNQYGNQ 718
L V AGT MARIWGRT CNFD +GRGRCQTGDC+GGL C GWG PPNTLAE+ALNQ+ N
Sbjct: 55 LDVAAGTKMARIWGRTNCNFDSSGRGRCQTGDCSGGLQCTGWGHPPNTLAEYALNQFNNL 114
Query: 717 DFYDISLVDGFNIPMDFFPLNGGCHKISCTADINGQCPNELRAPGGCNNPCTVYKTNEFC 538
DFYDISLVDGFNIPM+F P + CH+I CTADINGQCPN LRAPGGCNNPCTV++TN++C
Sbjct: 115 DFYDISLVDGFNIPMEFSPTSSNCHRILCTADINGQCPNVLRAPGGCNNPCTVFQTNQYC 174
Query: 537 CTNGQGTCGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCP 388
CTNGQG+C T++SRFFK RC D+YSYPQDDPTSTFTC +NY+VVFCP
Sbjct: 175 CTNGQGSCSDTDYSRFFKQRCPDAYSYPQDDPTSTFTC-TNTNYRVVFCP 223
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,066,484,550
Number of Sequences: 1393205
Number of extensions: 28825134
Number of successful extensions: 104996
Number of sequences better than 10.0: 493
Number of HSP's better than 10.0 without gapping: 85100
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 102193
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 64568047518
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)